Despite that leukemia in most cases is manifested as an expansion of haematopoietic progenitor cells of a defined lineage it is becoming increasingly clear that there are exceptions to this general concept. These include malignancies defined as acute leukemia of ambiguous lineage (ALAL) and can be manifested either as a bi-phenotypic disease with expansion of cells displaying combined expression of normally lineage restricted surface markers or bilinear leukemia, involving several lineages.Citation1 While this has been considered as a rare condition the use of targeted treatment of B-lineage acute leukemia (B-ALL) is beginning to unravel a surprising degree of plasticity in leukemia cells.
One of the most promising targeted treatments of refractory B-ALL is based on genetic manipulation of the patients own T-cells to make them reactive against CD19+ cells. This is achieved by retroviral transduction of the T-cells with a virus expressing a Chimeric Antigen Receptor (CAR). For B-ALL treatment the CAR carries a gene fusing an anti CD19 single chain variable fragment with signal transducing parts of T-cell activation receptors such as CD3 and/or CD28 to generate CAR-T cells. Despite that this approach presents challenges in managing acute symptoms caused by dramatic T-cell activation, it does in many cases represent an efficient treatment of severe cases of B-ALL.Citation2 However, in patients relapsing after treatment with CAR-T cells, a substantial fraction has developed CD19− leukemia. The importance of this is highlighted by the finding that 13 out of 20 children experiencing relapse after CAR-T cell treatment did so with CD19− leukemia cellsCitation3 suggesting that lineage instability in leukemia represent a clinically relevant problem.
In the paper by Somasundaram et al.Citation4 we report that using a mouse model for leukemia, we observed clonal conversion of B-ALL cells into either T or Myeloid cells. While cells converted to myeloid lineage underwent differentiation and did not generate leukemia, T-lineage converted cells generated a rapid in vivo expansion of immature T-lineage progenitors. This revealed that a leukemic state can be transferred between lymphoid lineages despite that the conversion process involve drastic changes in gene expression patterns as well as in the epigenetic landscape.Citation4 Our model is based on tumors developing in mice carrying heterozygote mutations in the Ebf1 and Pax5 genes encoding transcription factors critical for stable B-lineage identity.Citation5 Genetic alterations causing reduction in the functional dose of these proteins are common in human B-ALL with mutations or deletions in about 40% of the leukemia casesCitation6 and knock down of PAX5 or EBF1 levels in human leukemia cells resulted in increased lineage plasticity.Citation7 These findings suggest that the same regulatory networks that regulate normal B-cell development may be involved both in transformation as well as in lineage conversion processes, which further highlights the importance of these transcription factors in leukemia.
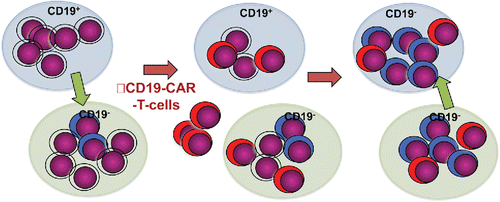
The conversion of B-ALL cells into T-lineage progenitors was dependent on an active Notch signaling which was achieved either by exposure to NOTCH ligand or as a result of ectopic expression of a constitutive active form of NOTCH1.Citation4 Hence, either genetic alterations causing activation of NOTCH signaling or the microenvironment may induce this conversion process. This opens up for the possibility that converted leukemia cells exist in specific microenvironments already at the onset of treatment (). The destruction of B-ALL cells by CD19 targeting CAR-T cells would then clear larger anatomic niches from CD19+ leukemia cells allowing for a rather rapid development of CD19− leukemia in the patient.
Figure 1. Schematic drawing representing the process by which a converted leukemic sub-clone escapes CAR-T cell treatment and expand in anatomic niches cleared of the CD19+ B-ALL cells. The purple cells represent B-ALL, blue cells represent converted cells and red cells represent CAR-T-cell. The blue and green areas represent different anatomic niches.
In our model system, we did not observe a malignant expansion of converted Myeloid cells despite that the conversion of human leukemia after CAR treatment often involves the formation of non-lymphoid leukemia.Citation3,7 Possible explanations to this would be that the genetic alterations underlying the transformation impact the ability of converted cells to adopt a malignant phenotype or that the experimental conversion process disrupt the oncogenic program.
In all, it is becoming increasingly clear that B-lineage leukemia cells are plastic in their nature and with increasing focus on investigations of targeted treatments these findings are likely to move from the laboratory into the clinic in the near future.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the Swedish Cancer Society, the Swedish Childhood Cancer Foundation, the Swedish Research Council, Knut and Alice Wallenbergs Foundation, Donation from Henry Hallberg and Linköping University.
References
- Manola KN. Cytogenetic abnormalities in acute leukaemia of ambiguous lineage: an overview. Br J Haematol 2013; 163:24-39; PMID:23888868; http://dx.doi.org/10.1111/bjh.12484
- Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M, Shah NN, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 2015; 385:517-28; PMID:25319501; http://dx.doi.org/10.1016/S0140-6736(14)61403-3
- Park JH, Geyer MB, Brentjens RJ. CD19-targeted CAR T-cell therapeutics for hematologic malignancies: interpreting clinical outcomes to date. Blood 2016; 127(26):3312-20; PMID:27207800; http://dx.doi.org/10.1182/blood-2016-02-629063
- Somasundaram R, Ahsberg J, Okuyama K, Ungerback J, Lilljebjorn H, Fioretos T, Strid T, Sigvardsson M. Clonal conversion of B lymphoid leukemia reveals cross-lineage transfer of malignant states. Genes Dev 2016; 30:2486-99; PMID:27913602; http://dx.doi.org/10.1101/gad.285536.116
- Somasundaram R, Prasad MA, Ungerback J, Sigvardsson M. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood 2015; 126:144-52; PMID:25990863; http://dx.doi.org/10.1182/blood-2014-12-575688
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007; 446:758-64; PMID:17344859; http://dx.doi.org/10.1038/nature05690
- Jacoby E, Nguyen SM, Fountaine TJ, Welp K, Gryder B, Qin H, Yang Y, Chien CD, Seif AE, Lei H, et al. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nature communications 2016; 7:12320; PMID:27460500; http://dx.doi.org/10.1038/ncomms12320
