Cell identity is controlled by complex gene regulatory networks that are subject to intricately orchestrated regulatory mechanisms.Citation1 Signaling pathways are the master choreographers of these networks, as they ensure execution of appropriate gene expression programs in accordance with key environmental cues, thereby providing critical spatial and temporal control of cell fate.Citation2 While signaling inputs directly modulate the activity of transcription factors, which serve as molecular determinants of cell identity, there is a growing appreciation that post-transcriptional mechanisms are an important effector arm of signaling cascades.
At the core of post-transcriptional regulation are numerous RNA-binding proteins (RBPs), which control the various steps in RNA's complex life cycles, from their maturation through their degradation.Citation3 RBPs act to facilitate the generation of cell fate diversity from similar primary transcriptomes, enforce transcriptional programs that maintain a given cell identity, and guide the timely transitions between different cell fates. As such, RBPs and their associated post-transcriptional mechanisms are well-positioned to be under the control of signaling pathways; yet, we have a rudimentary understanding of the integration between these two regulatory layers.
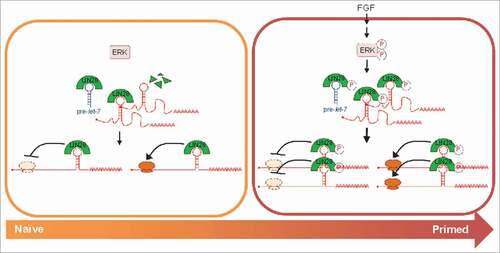
To gain insight into this problem, we recently explored the signaling-based control of LIN28, an ancient RBP that has emerged as a central regulator of cell identity in pluripotent stem cells.Citation4 Originally discovered in C. elegans as a regulator of developmental timing, LIN28 promotes the induction of pluripotency during somatic cell reprogramming and facilitates the transition from naïve to primed pluripotency.Citation5 At the molecular level, LIN28 is known to regulate two classes of RNA targets: the let-7 family of pro-differentiation microRNAs, whose biogenesis is blocked by LIN28, and select mRNAs, whose translation LIN28 can directly modulate.Citation4 Through phosphoproteomics analysis, we identified multiple phosphosites in LIN28 and found that one of them was targeted by MAPK/ERK, a critical signaling regulator of pluripotency.Citation6 This phosphorylation event stabilized the LIN28 protein, which had little impact on let-7 but enhanced its direct mRNA regulatory effects and pluripotency functions, overall establishing a link between signaling, post-transcriptional regulation, and cell fate control ().
Figure 1. Model of the coupling between signaling, post-transcriptional regulation, and cell fate control by the ERK-LIN28 axis. Left: In the absence of fibroblast growth factor (FGF), ERK is inactive, LIN28 is not phosphorylated and thus expressed at a lower level, whereby it can bind and inhibit the processing of let-7 precursors (pre-let-7) and engage with some of its direct mRNA targets to modulate their translation. Right: Upon FGF stimulation, ERK is activated and phosphorylates LIN28, which increases LIN28s protein levels, allowing it to engage with more mRNA targets and enhancing its effect on translation. As indicated on the bottom, this mechanism facilitates the transition from naïve to primed pluripotency.
These findings provide an example of what is likely a widespread regulatory mechanism, whereby signaling pathways modulate RBPs and their associated regulatory networks to influence cell identity. Such an integration offers several unique opportunities for robust cell fate control. First, post-transcriptional regulation can act faster than transcriptional control, a feature that is well-aligned with the rapid-response nature of signal transduction and the critical role of timing in developmental processes. Second, since signaling is driven by environmental cues, its coupling to RBPs enables direct influence of the niche on the existing cellular transcriptome, communication that is essential to cell fate specification. Third, it allows for multi-faceted control of RNA molecules by affecting properties other than their expression levels alone, thus providing additional mechanisms for signaling pathways to impact cell identity.
Direct signaling input into an RBP – such as phosphorylation – can affect various RBP properties. In the case of LIN28, we uncovered an effect on its stability, which fundamentally alters the stoichiometry of the RBP relative to its targets. However, an RBP's subcellular localization or association with critical protein co-factors and RNA targets may also be influenced through changes in the RBP's binding affinity or availability of relevant domains. And, for RBPs that enzymatically alter RNAs, their catalytic activity might also be modulated. At the same time, as already noted, an RBP can regulate various aspects of RNA metabolism, including processing, localization, translation, and degradation. Altogether, the combination of possible effects on RBP properties and downstream effects on its cognate RNA targets generates a diverse array of outputs that can be controlled by signaling pathways.
Building a more complete picture of these regulatory relationships will require a systematic interrogation of the RNA-binding proteome. While LIN28 provides a valuable lens into this biology, there are over 500 RBPs associated with pluripotency alone, most of which have not been functionally characterized.Citation7 Hence, a first step would be to define their functions through a combination of genetic and biochemical approaches. Pluripotent stem cells provide a good starting platform, which could be expanded to differentiated cell types, since RBP function is highly context-dependent (as it is restricted by the available transcriptome). It would further be informative to analyze transitions between cell fates, as RBPs may be particularly relevant in these settings (by enforcing transcriptome switches). And, while rare, systems that rely entirely on post-transcriptional control, such as the earliest stages of embryogenesis or reticulocyte maturation, could provide powerful insights into core regulatory principles. Once a functional understanding is gained, the connection to signaling pathways can be explored. This can be done in a signaling-centric manner, by analyzing changes in an RBP and its targets upon pathway perturbation, or in an RBP-centric manner, such as the phosphoproteomic analysis performed for LIN28. The accumulating public data from global transcriptomic and proteomic studies may offer a useful resource in that respect.
Ultimately, such an approach can bring us closer to charting RBP-based post-transcriptional regulatory networks and yield a more comprehensive understanding of the molecular foundations of cell identity.
Disclosure of potential conflicts of interest
G.Q.D. holds equity and receives consulting fees from 28/7 Therapeutics, a biotechnology company seeking to develop small molecule inhibitors of the LIN28/let-7 pathway.
References
- Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. CellNet: Network biology applied to stem cell engineering. Cell 2014; 158:903-15; PMID:25126793; http://dx.doi.org/10.1016/j.cell.2014.07.020
- Perrimon N, Pitsouli C, Shilo BZ. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb Perspect Biol 2012; 4:a005975; PMID:22855721; http://dx.doi.org/10.1101/cshperspect.a005975
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008; 582:1977-86; PMID:18342629; http://dx.doi.org/10.1016/j.febslet.2008.03.004
- Shyh-Chang N, Daley GQ. Lin28: Primal regulator of growth and metabolism in stem cells. Cell Stem Cell 2013; 12:395-406; PMID:23561442; http://dx.doi.org/10.1016/j.stem.2013.03.005
- Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D, Xia Q, Seligson M, et al. LIN28 regulates stem cell metabolism and conversion to primed pluripotency.Cell Stem Cell 2016; 19:66-80; PMID:27320042; http://dx.doi.org/10.1016/j.stem.2016.05.009
- Tsanov KM, Pearson DS, Wu Z, Han A, Triboulet R, Seligson MT, Powers JT, Osborne JK, Kane S, Gygi SP, et al. LIN28 phosphorylation by MAPK/ERK couples signalling to the post-transcriptional control of pluripotency.Nat Cell Biol 2017; 19:60-7; PMID:27992407; http://dx.doi.org/10.1038/ncb3453
- Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 2013; 20:1122-30; PMID:23912277; http://dx.doi.org/10.1038/nsmb.2638
