MicroRNAs (miRNA) are a class of small non-coding RNAs, which control the expression of protein coding genes via translational repression and/or mRNA degradation. They regulate all biologic processes through their function in the networking and fine-tuning of gene expression, facilitating rapid switches during developmental programs or acting as guardians of established cellular fates. Dysregulation of the miRNA biogenesis pathway, via the deletion of their effector protein genes i.e Dgcr8 or Dicer, during mouse early development leads to embryonic post-implantation lethality (reviewed in ref. Citation1). Likewise, in vitro studies demonstrated that mouse Embryonic Stem Cells (mESCs) depleted for Dgcr8 or Dicer genes present deleterious effects in proliferation, cell cycle progression and differentiation.Citation2,3 Interestingly, the Dgcr8 and Dicer-deficient mESCs were generated in different genetic backgrounds and presented subtle differences in their phenotypes,Citation1 which were mainly attributed to the physiological role of DICER-dependent endogenous small RNAs in mESCs.Citation2
Recent advances in genome engineering allowed us to mimic the original deletions of Dgcr8 and Dicer genes in mESCs with the same genetic background using paired CRISPR-Cas9 technology.Citation4,5 An in-depth characterization of the generated mutant cells reproduced previous findings and revealed new phenotypes. In addition to their aforementioned defects, Dgcr8 and Dicer mutant mESCs presented a reinforcement of the pluripotency network and an impaired exit from the pluripotent state.Citation4,5
From these experiments, we concluded that a correct post-translational modification of the protein is essential for stem cells commitment, independently of the role of DGCR8 in miRNA biogenesis. Indeed, recent studies revealed non-canonical functions for RNA interference (RNAi) proteins in several biological processes.Citation1
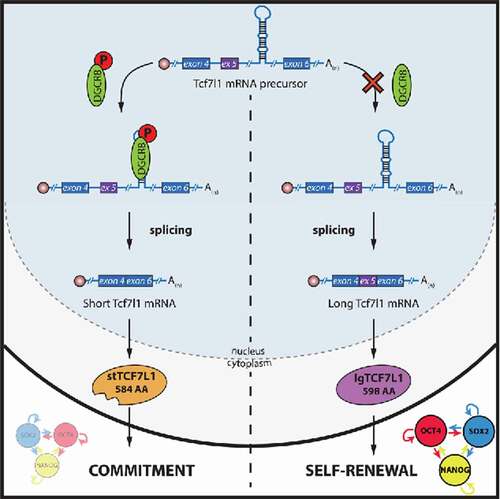
To identify the underlying mechanisms responsible for the blockade of pluripotency exit in Dgcr8 mutant mESCs, we used an integrative analysis of genomic data sets. Intersection of the differential gene expression in Dgcr8-deficient versus WT mESCs, the transcripts bound by DGCR8 and the genes required to exit from pluripotency, demonstrated that a correct phosphorylation of DGCR8 protein was essential for its binding to the stem cell-specific long Tcf7l1 mRNA isoform,Citation4 facilitating, subsequently, its own proper splicing into a short TCF7L1 protein and allowing the repression of pluripotency-associated genes ().
Figure 1. DGCR8 regulates mESC fate. During mESC commitment, phosphorylated DGCR8 binds to a stem loop structure present in Tcf7l1 mRNA, facilitating its splicing into a short Tfc7l1 isoform, which will subsequently promote differentiation. Conversely, when DGCR8 is not present or hypophosphorylated, splicing is impaired and a longer TCF7L1 protein is produced, resulting in the activation of pluripotency-associated genes or hampering the dissolution of the pluripotency network.
Although our work highlights a new non-canonical function for DGCR8 in mESCs, it leaves some important questions unanswered. The post-translationally impaired DGCR8 variant used in our study contains 23-phosphosite-mutant residues localized in the unstructured region of DGCR8 that do not impair the interaction with its miRNA biogenesis partner DROSHA neither miRNA production.Citation4,6 We hypothesize that these 23 amino acid mutations (Serine or Threonine changed to Alanine and Valine) affect the charge of the protein and therefore change the affinity of DGCR8 for specific mRNAs. Nevertheless, it is still unclear which phospho-sites are responsible for the correct binding of DGCR8 to the Tcf7l1 mRNA. Complementation of the Dgcr8-deficient mESCs with different phospho-mutant DGCR8 variants and assessing the exit from pluripotency as well as the splicing of Tcf7l1 mRNA should reveal the key phospho-residues of DGCR8 protein. Furthermore, which kinases phosphorylate DGCR8 in mESCs and how they are regulated during early mammalian development remain open questions.
Importantly, DROSHA is able to interact with both phosphorylated and non-phosphorylated DGCR8 proteins allowing the production of microRNAs.Citation4 However, it is possible that the change in the post-translational status of DGCR8 impacts its interaction with other proteins. The analysis of the 2 DGCR8 variant interactomes in mESCs may reveal specific stem cell interactors of DGCR8, which could also be directly involved in the splicing of the Tcf7l1 mRNA.
In addition, it is legitimated to ask whether other mRNA splicing events might be regulated by DGCR8 and could be identified. Although the binding of DGCR8 might not always lead to changes in relative mRNA expression or alternative splicing, but rather as a scanning function for the target mRNAs. Two other important questions might be raised from our study: How general is this new mechanism in the control of RNAs fate? Is this mechanism conserved in other cell types or organisms? To answer these questions several biochemical approaches might be considered in ESCs or progenitor cells from other species, i.e RIP-seq (RNA-immunoprecipitation followed by high throughput sequencing) or HIT-CLIP (High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation).
Finally, this novel non-canonical function of DGCR8 points to a reinterpretation of the phenotypes previously observed for the other RNAi members in mESCs (e.g. DROSHA, DICER and AGO proteins) that were mainly attributed to the deficiency of miRNAs.Citation1 Of note, deletion of the genomic region (22q11.2) in DiGeorge syndrome patients involves more than 30 genes, including Dgcr8. Mouse models lacking this genomic region showed minor alteration of miRNAs, but strong behavioral and cognitive deficits and cardiac abnormalities.Citation7 Now, that new evidences revealed an important role of DGCR8 in the regulation of splicing and in the exit from pluripotency, it would be of great interest to determine how important these functions are for the origin and development of the disease.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Bodak M, Cirera-Salinas D, Luitz J, Ciaudo C. The role of RNA interference in stem cell biology: Beyond the mutant phenotypes. J Mol Biol 2017; in Press; PMID:28118980; http://dx.doi.org/10.1016/j.jmb.2017.01.014
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 2007; 39:380-5; PMID:17259983; http://dx.doi.org/10.1038/ng1969
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. PNAS 2005; 102:12135-40; PMID:16099834; http://dx.doi.org/10.1073/pnas.0505479102
- Cirera-Salinas D, Yu J, Bodak M, Ngondo RP, Herbert KM, Ciaudo C. Non-canonical function of DGCR8 controls mESCs exit from pluripotency. J Cell Biol 2017; 216:355-66; PMID:28100686
- Bodak M, Cirera-Salinas D, Yu J, Ngondo RP, Ciaudo C. Dicer, a new regulator of pluripotency exit and LINE-1 elements in mouse embryonic stem cells. FEBS Open Bio 2017; 7:204-20; PMID:28174687; http://dx.doi.org/10.1002/2211-5463.12174
- Herbert KM, Pimienta G, DeGregorio SJ, Alexandrov A, Steitz JA. Phosphorylation of DGCR8 increases its intracellular stability and induces a progrowth miRNA profile. Cell Rep 2013; 5:1070-81; PMID:24239349; http://dx.doi.org/10.1016/j.celrep.2013.10.017
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills A A, Karayiorgou M, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet 2008; 40:751-60; PMID:18469815; http://dx.doi.org/10.1038/ng.138
