ABSTRACT
Defects in apoptotic pathway contribute to development and progression of oral cancer. Survivin, a member of the inhibitors of apoptosis protein (IAP) family, is increased in many types of cancers. However, it is unclear whether increased survivin is associated with oral squamous cell carcinomas (OSCC), and what mechanisms may involve in. In this study, we examined survivin expression in OSCC compared with normal oral tissues via immunohistochemical staining. The results showed that, not only total survivin is increased in OSCCs, but also the subcellular location of survivin is changed in OSCCs compared with normal oral tissues. In most of normal oral tissues, survivin staining was either negative, or cytoplasmic positive/nuclear negative; whereas in most of OSCC tissues, survivin staining was nuclear positive. Statistic analysis indicates that nuclear survivin, rather than total or cytoplasmic one, correlates with tumor TNM stage and differentiation grade. Consistently, in vitro analysis showed that survivin is in cytoplasm in normal human oral kinotinocyte (HOK) cells; whereas it is in nucleus in OSCC HN6 cells. Importantly, treatment of HOK cells with HDAC inhibitor Trichostatin A (TSA) induces survivin acetylation and promotes its nuclear localization. Moreover, nuclear survivin in OSCC cells was acetylated at K129 in its C-terminal, suggesting that the acetylation is important for nuclear location of survivin. Our study demonstrates that it is nuclear survivin, rather than total or cytoplasmic one, associates with TNM stage and tumor grade of OSCC. Thus, we propose nuclear survivin as a prognostic marker for the progression of OSCC.
Introduction
Oral cancer has been an important component of the worldwide burden of cancer with about 300,000 new cases every year.Citation1 Oral squamous cell carcinoma (OSCC) accounts for 95% of oral cancers. Although many efforts have been made to improve the diagnosis and treatment of OSCC patients, the prognosis of the OSCC is still poor and the overall survival rate is less than 50% in 5 y.Citation2 One of the primary reasons for the poor prognosis in OSCC is lack of unique molecular tumor markers to assess the risk and prognosis. Although numerous tumor markers and risk factors of OSCC have been proposed, the prognostic value of these markers is still of controversy.Citation3
Programmed cell death via apoptosis serves as a natural barrier to cancer development.Citation4 Tumor cells evolve a variety of strategies to limit or circumvent apoptosis. Survivin is a member of the inhibitors of apoptosis protein (IAP) familyCitation5 that plays an important role in suppression of apoptosis by inhibiting the activity of caspases.Citation6-8 Survivin expression is detectable at high levels in embryonic tissues, but is at low or non-detectable levels in normal adult tissues with exception of thymus, basal colonic epithelium, endothelial cells, and neural stem cells.Citation9,10 Importantly, survivin is upregulated in most types of human cancers, including lung, breast, colon, stomach, esophagus, pancreas, bladder, uterus, ovary, liver carcinomas, neuroblastoma, glioma, melanoma, soft tissue sarcoma, leukemias, and high-grade non-Hodgkin lymphoma.Citation11-13 The high levels of survivin have been linked to its anti-apoptotic activity development in tumors. In addition, it has also been described that survivin in different subcellular location, cytoplasm or nucleus, have distinct functions, such as inhibition of apoptosis or regulation of cell proliferation. However, there are controversy reports regarding the biological roles of survivin in different subcellular locations. Some reports suggested that nuclear survivin was involved in promoting cell proliferation; whereas cytoplasmic survivin is involved in controlling cell survival.Citation14,15 Some reports showed that cytoplasmic survivin was associated with a poor prognosis in cancer patients,Citation16,17 whereas other reports gave the opposite conclusions regarding to the role of nuclear survivin.Citation18 Taken together, the biological roles or prognostic values of distinct subcellular localized survivin in OSCC are unclear, and the mechanism involved in regulation of subcellular localization of survivin remains elusive.
In this study, we examined survivin expression in 90 paired samples of OSCCs and adjacent normal oral tissues by immunohistochemical staining (IHC). We found that nuclear survivin, rather than the total or cytoplasmic one, was associated with TNM stage and tumor grade of OSCC. Moreover, we showed that acetylation of residue K129 in the C-terminal is involved in regulation of nuclear translocation of survivin. We proposed that nuclear survivin is a prognostic molecular marker for OSCCs.
Results
Patient characteristics
We collected 90 paired OSCC and adjacent normal oral tissue samples. Among them, 35 cases (38.8%) were well differentiated, 46 (51.1%) were moderately differentiated, and 9 (10%) were poorly differentiated. The classification of cancer stage of samples was based on TNM, including 22 cases in stage I, 24 in stage II, 23 in stage III, and 21 in stage IV. There were no cases of distant metastasis. Of 40 leukoplakia patients, Twenty (50%) were men and 20 were women, with a mean age of 59 y (SD 11.6, range 32–80 years).The characters of OSCC patients analyzed in the study were summarized in .
Table 1. The baseline characteristics of OSCC patients include in the study.
Survivin mRNA expression is significantly upregulated in OSCC and predicates a poor prognosis
To compare the mRNA levels between normal oral tissues and OSCCs, we searched the data from Oncomine (www.oncomine.com). A data set that includes survivin gene expression profiling was available from the report of Peng et al..Citation19 We found that survivin mRNA in OSCC was significantly higher than that in normal oral tissue (). Because the survival data were not available for this study, we further selected one independent study on head and neck cancer from TCGA (TCGA, Nature 2015). We performed analysis by using the cBio Cancer Genomics Portal (http://cbioportal.org).Citation20 Specifically, the expression levels of survivin gene were divided into 2 groups, high expression and low expression groups. By analysis of the overall survival based on survivin mRNA expression, we found that patients with high survivin levels had a significantly shorter median overall survival than patients with low survivin levels (). Thus, overexpression of survivin predicates a poor prognosis and may play a critical role in OSCC.
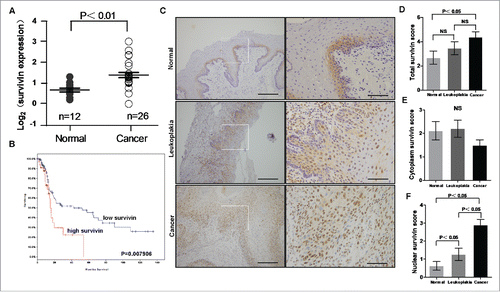
Figure 1. Survivin is upregulated in OSCC. (A) Oncomine-derived data analysis of survivin mRNA expression in normal oral tissue and OSCC. (Kruskal-Wallis test, P < 0.01; normal oral n = 12, OSCC n = 26) (B) Patient survival obtained from publicly available microarray data was analyzed based on survivin mRNA expression level. (C) Representative images of survivin expression in normal oral tissues, leukoplakias and OSCCs via immunohistochemical (IHC) staining. Scale bar: 50 um. (D) IHC scores of survivin expression in normal, leukoplakia and OSCC tissues.
Nuclear survivin is associated with OSCC tissue samples and predicts the aggressive clinicopathological characters in OSCC patients
To further determine the levels of survivin in oral tissues, we examined survivin expression in 90 paired primary OSCCs and adjacent normal oral tissue and 40 leukoplakia patients by immunohistochemistry (IHC). We found that the total survivin level in OSCCs was significantly higher than that in normal oral tissues, but there was no significant difference between normal and leukoplakia or between leukoplakia and OSCCs. (). We also noticed that the subcellular locations of survivin are different between normal oral tissues and OSCCs. Then, we quantified the portions of cytoplasmic and nuclear survivin in the 90 paired oral normal and OSCC and 40 leukoplakia samples. The result indicates that although the total survivin () is higher in OSCCs than that in normal oral tissues, the cytoplasmic survivin was actually slightly lower in OSCC than that in normal oral tissues (P = 0.154, no statistical significance) (). Importantly, we found that the nuclear survivin is dramatically higher in OSCC tissues than that in leukoplakias and normal oral tissues (). This pattern suggests that nuclear survivin is associated with the progression in OSCCs.
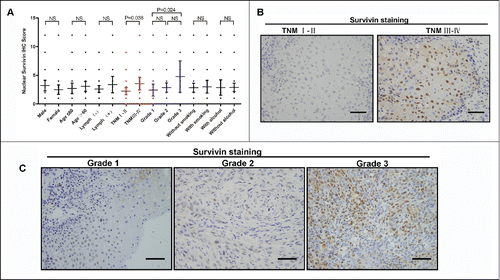
To determine the clinical relevance of nuclear survivin in the progression of OSCC, we quantified the IHC scores of total, cytoplasm, and nuclear survivin respectively, and compared them with the clinico-pathological characteristics. The results showed that the total and cytoplasm survivin is not associated with the clinicopathological characters (data not shown). But the nuclear survivin in OSCC tissues is significantly correlated with aggressive clinicopathological characters, such as TNM stage and tumor grade (). Tumors with late TNM stage and poor differentiation grade show a relative high nuclear survivin expression ().
Figure 2. Up-regulated nuclear survivin expression predicts the aggressive clinicopathological characteristics in OSCC patients. (A) Different nuclear survivin expression levels in 90 OSCC patients based on clinicopathological characteristics. (NS, P > 0.05) (B) Representative IHC images of nuclear survivin in OSCCs with different TNM stage. Scale bar: 50 um (C) Representative IHC images of nuclear survivin in OSCCs with different differentiation grades. Scale bar: 50 um.
Survivin is localized in cytoplasm in normal oral keratinocyte cells and in nucleus in oral cancer cells
To determine the expression profiles of survivin in oral epithelial cells and oral cancer cells, we performed immunoblot analysis on survivin levels in one normal human oral keratinocyte (HOK) cell line and 7 OSCC-derived cell lines (). The result indicates that survivin is relatively low in HOK cells, whereas the levels of survivin were relatively higher in most of 7 OSCC cell lines examined. This observation suggests that although survivin is expressed at a low level in normal oral keratinocyte cells, its expression is frequently upregulated in OSCC cells.
Figure 3. The expression and subcellular location of survivin in normal HOK and oral cancer cell lines. (A) Western blotting analysis of survivin in human oral keratinocytes (HOK) cells, and 7 OSCC lines. (B) The subcellular location of survivin in HOK and HN6 cell lines. Immunofluorescence with antibody to survivin was used to localize survivin. The nuclei were stained with DAPI. (C-D) Cellular fractionation and immunoblot were performed to determine the cytoplasm/nuclear survivin ratio in HOK and HN6 cells.
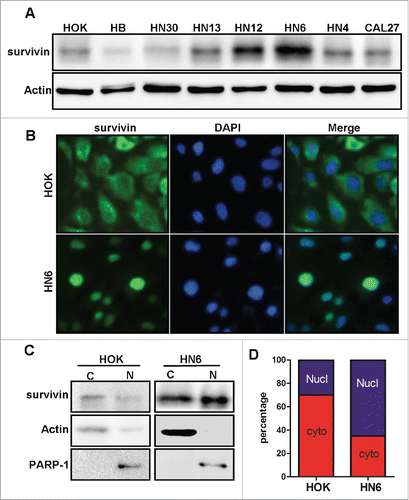
To determine the subcellular locations of survivin in vitro, we examined via immunofluorescence (IF) staining the subcellular location of survivin in HOK cells and HN6 cells, an OSCC-derived cell line. We found that survivin expression in HOK cells is mostly in cytoplasm (); while survivin in HN6 cell is mainly in nucleus (). To further validate the results, we also performed cell fractionation and immunoblot for survivin in different subcellular portions. The results showed that the portion of nucleus survivin was significant enhanced in HN6 cells than in HOK cells (). These results are consistent with IHC analysis on clinical tissue samples, suggesting that nuclear survivin, rather than cytoplasmic one, correlates with malignant phenotype in oral cancer tissues.
Acetylation at K129 of survivin is involved in induction of nuclear translocation of survivin in OSCC cells
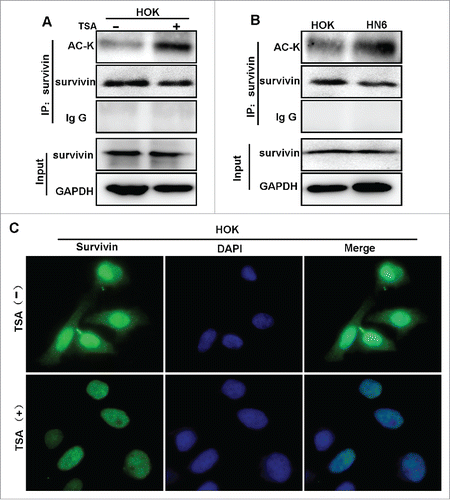
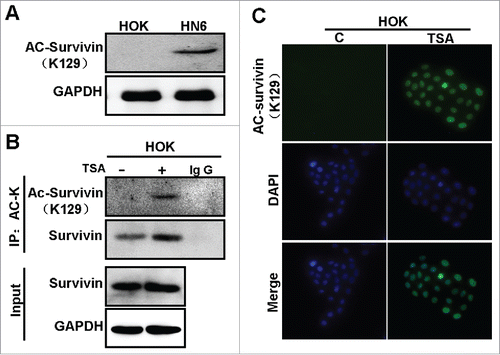
To determine which signal pathway may involve in induction of nuclear translocation of survivin in OSCCs, we examined the effects on the subcellular location of survivin in HN6 oral cancer cells by treatment with different chemical inhibitors of different pathways, including STAT3 inhibitor (Stattic), PI3K inhibitor (LY294002), NF-βB inhibitor (BAY11–7082), and β-catenin/TCF inhibitor (FH535). We performed cell fractionation and immunoblot for survivin in different subcellular portions. However, none of these inhibitors affects the subcellular location of survivin (data not shown). Previously, Wang et al. demonstrated that survivin can be acetylated in a growth-factor-dependent manner by the histone acetyl-transferase (HAT), CREB-binding protein (CBP) in breast cancer cells.Citation21 Moreover, CBP-dependent acetylation at lysine 129 (Lys-129) of survivin is suggested to promote its homodimerization and stability in nucleus. Thus, we hypothesized that acetylation of survivin is involved in regulation of subcellular localization of survivin in OSCC. To determine if survivin could be acetylated in OSCC, we first treated HOK cells with the histone deacetylase inhibitor Trichostatin (TSA), then analyzed acetylated survivin by IP-Western. We found that acetylated survivin was detected by pan-acetylated-lysine (pan-ACK) following IP (). Next, we examined acetylated survivin in HOK and HN6 cells by IP-Western. Interestingly, more acetylated survivin was detected in HN6 cells than in HOK cells (). To validate the observation, we examined the subcellular location of survivin in HOK cells following treatment with TSA by IF analysis. As shown in , TSA treatment drastically reduced the cytoplasmic survivin, and simultaneously increased the nuclear survivin. Taken together, these results suggest that acetylation of survivin is involved in promotion of nuclear translocation of survivin.
Figure 4. Acetylation promotes survivin nuclear localization in OSCC cells. (A) Cell lysates of HOK with or without treatment of TSA were subjected to immunoprecipitation by anti-survivin, and then subjected to Westernblot with anti-pan-acetylation (AC-K) or anti-full-length survivin. (B) Cell lysates of HOK and HN6 cells were subjected to IP with anti-survivin, and to Westernblot with anti-pan-acetylation (AC-K) or anti-full-length survivin. (C) HOK cells treated with or without TSA were analyzed via IF staining with anti-survivin.
To determine if nuclear survivin is acetylated at K129 in OSCC, we applied antibody specific to acetylated K129 in analysis of acetylated survivin in HOK and HN6 cells. The results showed that K129-acetylated survivin is readily detected in HN6 cells, but not in HOK cells (). Then, we treated HOK cells with or without TSA and immunoprecipitated with a pan-acetylated-lysine (pan-ACK) antibody. We found that K129-acetylated survivin was increased in HOK cells treated with TSA. We further examined the localization of K129-acetylated survivin by immunofluorescence. As shown in , K129-acetylated survivin was not detected in HOK cells in the absence of TSA. However, in the presence of TSA, the K129-acetylated survivin was detected and highly enriched in the nucleus (C). These results suggest that nuclear survivin is acetylated at K129, and acetylation is important in regulation of nuclear translocation of survivin in OSCC cells.
Figure 5. Acetylation at lysine 129 promotes survivin nuclear localization. (A) Cell lysates of HOK and HN6 cells were subjected to Westernblot with anti-129 K acetylated survivin. (B) Cell lysates of HOK cells treated with or without TSA were subjected to IP with anti-pan-acetylation (AC-K), and were then subjected to Westernblot with anti-129 K acetylated survivin or anti-full-length survivin. (C) IF of HOK cells treated with or without TSA and immunostained with anti-129 K acetylated survivin.
Discussion
In this study, we showed that survivin expression is significantly lower in normal oral tissues than in tumor tissues. Importantly, survivin in normal oral epithelium was mainly localized in cytoplasm; whereas survivin in OSCCs was mainly, in nucleus. Moreover, we found that nuclear survivin was associated with tumor TNM stage and tumor grade. And finally, we showed that acetylation at K129 of survivin is involved in regulation of nuclear localization of survivin in OSCC cells. Taken together, our study provides a biochemical and biological link between the pathological features and subcellular location of survivin in OSCCs.
Generally, survivin is only expressed during embryonic and fetal development as a means to regulate proper cell division and growth. It is undetectable in most terminally differentiated normal tissues.Citation11 However, survivin is constitutively expressed in some normal adult tissues, such as haematopoietic stem cells,Citation22 thymocytes,Citation12 melanocytes,Citation11 gastric mucosa and colonic epithelium.Citation23 Although survivin was reported to be negative in normal mucosa specimens from the upper aero-digestive tract,Citation14 our analysis shows that survivin is positive, although weakly, in cultured non-neoplastic cells of oral mucosa.
It has been reported that survivin expression level is associated with a poor outcome in cancer patients. However, this conclusion was based on the analysis of mRNA level or total, including cytoplasmic, survivin. Citation24-26 In a previous study on oral cancers, evaluation of the prognostic significance was based on the total survivin.Citation27 However, the influence of subcellular location of the protein did not take into account. Thus, it remains controversial regarding the prognostic value of survivin expression cells in OSCCs. In this study, survivin expression was analyzed in both cytoplasm and nucleus, in OSCC cell lines, and in the samples of normal oral and OSCC tissues. In particular, our analysis was focus on the subcellular location of survivin in regarding the prognosis of OSCCs.
Clinically, cancer stages are based on TNM staging system, which reflects tumor growth through proliferation (T-score) and lymph node metastasis (N-score). Histological tumor grade has been reported to be predictive for the recurrence in patients in an early stage of OSCCs.Citation28 The 5-year survival rates of OSCC patients in stage III and IV are 20–43%.Citation29 In this study, we found that nuclear survivin was correlated with TNM stage and histological tumor grade. This suggests that nuclear survivin is as a promising molecular marker for OSCC progression and prognosis. Consistent with our observation, nuclear survivin, as by IHC staining, was also implicated in association with the prognosis of gastric, breast, bladder cancers, osteosarcoma,Citation30-33 NSCLCsCitation16 and in esophageal squamous cell cancer.Citation34
Survivin exists in 2 subcellular pools (cytoplasmic and nuclear), however the roles of subcellular localization of survivin expression in malignant cells is still controversial. Fortugno et al.Citation35reported immunochemically distinct nuclear and cytoplasmic pools of survivin involved in regulation of mitotic spindle microtubules formation and function. Mahot ka et al.Citation36 described that survivins of different splice variants have differential subcellular localization and functions. It has been shown that cytoplasmic survivin is mainly non-anti-apoptotic survivin-2B; whereas nuclear survivin is mainly anti-apoptotic isoform survivin-βEx3. De MariaCitation37 reported that both survivin-β Ex3, and survivin-2B were significantly increased in OSCCs, whereas survivin-3B was only slightly (not significant) increased. In this study, the subtypes of survivin have not been examined. Thus, the relationship between the subtypes of survivin expression and tumor progression remain unknown, which remains to be investigated in future.
Post-translational modifications (PTMs) are the important mechanism in regulation of survivin. Several reports highlight the role of PTMs (phosphorylation, acetylation and ubiquitination) in the regulation of survivin functions. Through phosphorylation, CDC-cyclin-B1 kinase (Cdk1), aurora-B kinase, Polo-like kinase 1 (Plk1) and casein kinase 2 (CK2) can regulate survivin mitotic function via phosphorylation on threonine 34, threonine 117, serine 20 and threonine 48Citation38-41 or regulate apoptosis via phosphorylation on threonines 34 and 48.Citation42 In addition, other types of PTMs are also implicated in regulation of survivin localization and its functions. For example, several lysine residues (23, 90, 110, 112, 115, 120, 121, 122, 129 and 130) in survivin are potential targets of the histone acetyltransferase CREB (cAMP response element-binding protein)-binding protein (CBP).Citation43 Acetylation at lysine 129 is suggested to promote homodimerization of survivin, which leads to localization in the nucleus; whereas deacetylation at lysine 129 promotes the formation of monomers that heterodimerizes with chromosome region maintenance 1 (CRM1), leading to nuclear export.Citation21 However, the biological function and clinical relevance of this modification was unclear. In this study, we showed that acetylation at K129 contribute to nucleus localization of survivin in OSCC. Moreover, nuclear survivin, rather than total or cytoplasmic one, is associated with TNM stage and tumor grade of OSCC. Thus, post-translational modification by acetylation at Lys-129 of survivin facilitates its nuclear localization, and promotes tumor progression in OSCC.
There are still some limitations in this study. Firstly, the sample size (90 cases) in this study may be not big enough to support a powerful conclusion. Second, the detailed mechanisms regarding the regulation of survivin nuclear localization by acetylation is not fully understood, which requires further characterization. Third, the survival data of the OSCC tissue samples used in IHC analysis in study is currently unavailable, which requires more time to collect the data on the following-up study. Nevertheless, our study indicates that, it is the nuclear survivin, rather than the total or cytoplasmic one, that has a prognostic value for OSCCs. We propose nuclear survivin as a biomarker for diagnosis, and as a target for therapy of OSCCs.
Materials and methods
Patients and tissue samples
Tissue specimens used in this study comprised 90 surgically resected oral squamous cell carcinomas and 40 leukoplakia samples collected under a hospital Review Borad protocol and archived as formalin-fixed paraffin-embedded (FFPE) specimens in the Ninth People's Hospital (Shanghai, China) from 2007 to 2011. The 90 primary OSCC patients enrolled in this study were without prior radiotherapy or chemotherapy. The age of these patients ranged from 18 to 92 y with an average 59.8 y. The sites of the primary carcinoma were tongue (n = 44), buccal mucosa (n = 14), floor of mouth (n = 11), gingiva (n = 14), and soft palate (n = 7). Histopathologic diagnosis of each neoplastic tissue was performed according to the World Health Organization criteria by the Department of Oral Pathology, Ninth People's Hospital of Shanghai. Clinicopathologic staging was determined by the TNM classification of the International Union against Cancer. Our study was approved by the ethical committee of the Ninth People's Hospital of Shanghai, China. Ninety paired clinically normal oral mucosa specimens, distant from tumor sites, were obtained at least 2 cm away from the edge of tumor mass, with best efforts of avoiding contamination of the tumor cells and conformed by 2 experienced pathologists.
Cell culture and reagents
Human oral keratinocyte (HOK) was purchased from Chinese Beijing North (Beijing, China), a Biotechnology Research Institute, as described before.Citation44 OSCC-derived cell lines HN4, HN6, HN12, HN13, HN30, HB was kindly provided by the University of Maryland, School of Dentistry. CAL27 cell line was purchased from the American Type Culture Collection (ATCC). HOK cells were cultured in RPMI-1640 Medium (Invitrogen) supplemented with 15% fetal bovine serum (FBS). Other cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum, 1% glutamine, and 1% penicillin–streptomycin. All cells were maintained in a humidified atmosphere of 5% CO2 at 37°C. Antibodies against PARP-1 were from Santa Cruz Biotechnology. Antibodies for survivin, AC-K, Actin, GAPDH were from Cell Signaling Technology Inc. Antibodies for anti-acetylatedK129 survivin antibodies were from Novus Biologicals. HRP-conjugated secondary antibodies were from eBioScience.
Immunohistochemistry and evaluation of staining
Immunohistochemistry was performed as described previously.Citation45 Immunohistochemical staining was performed on the sections (3-μm thickness) of paraffin-embedded specimens with rabbit anti-survivin (RB-9245-R7, NeoMarkers). Briefly, the sections were dewaxed in xylene and hydrated with graded ethanol. Then antigen retrieval was performed using 0.01 mM citrate buffer (pH 6.0) pressure-cooking, and endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 10 minutes at room temperature. The slides were incubated with anti-survivin (Ready-to-use) in a moist chamber for overnight at 4°C. Upon incubation with the primary antibody, the specimens were washed 3 times in PBS and visualized using 3,3′-diaminobenzidine detection kit (Dako Cytomation, Denmark). Samples were then counterstained with hematoxylin, a blue nuclear stain. Tumor cells were considered survivin-positive if the immunoreactivity was observed in the nucleus and/or cytoplasm. The mean percentage of positive tumor cells was determined at least 5 random fields at ×400 magnification in each section. The percentage of positive tumor cells was assigned to one of the following categories: 0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. The intensity of survivin immunostaining was scored as follows: (a) weak, 1+; (b) moderate, 2+; and (c) intense, 3+. The percentage of positive tumor cells and the staining intensity were multiplied to produce a weighted score for each case. Cases with a weighted survivin score≧2 were considered to be positive. These judgments were made by 2 independent pathologists, neither of whom had knowledge or information pertaining to the patients' clinical status.
Immunofluorescence
Immunofluorescence was performed as described previously.Citation46,47 Cultured cells were rinsed 3 times with PBS and fixed with 3.7% formaldehyde and permeabilized with 0.1% Triton X-100. After blocking in 1% BSA for 1 hour, the cells were incubated in a moist chamber for overnight at 4°C with anti-survivin (NOVUS, NB500–201, dilution 1:300), then washed and incubated for 1 hour with Alexa Fluor 488 donkey anti-rabbit IgG (H+L) antibody (Invitrogen, A21206, dilution 1:400) at room temperature in dark. The cells were washed 3 times with PBS containing 0.02% Tween20 and mounted in aqueous mounting medium containing 0.5 mg/ml 40–6-diamidino-2-phenylindole to stain the nuclei. Cells were examined under a fluorescence microscope at 400× magnification.
Cellular fractionation
Cellular fractionation was performed as described previously.Citation48 Briefly, cells were pelleted and resuspended in 300 µl of buffer A (50 mM NaCl, 10 mM HEPES pH 8, 500 mM sucrose, 1 mM EDTA, 0.5 mM Spermidine, 0.15 mM Spermine, 0.2% Triton X-100) containing β-mercaptoethanol and the protease inhibitors PMSF, leupeptin, aprotinin and pepstatin. After 15 min on ice and centrifugation at 9000 rpm for 15 minutes, the supernatant (cytoplasmic fraction) was collected and stored. The pellet was washed with 200 µl Buffer B (50 mM NaCl, 10 mM HEPES pH 8, 25% glycerol, 0.1 mM EDTA, 0.5 mM Spermidine, 0.15 mM Spermine) and then resuspended in 100 µl buffer C (350 mM NaCl, 10 mM HEPES, 25% glycerol, 0.1 mM EDTA, 0.5 mM Spermidine, 0.15 mM Spermine). After centrifugation at the maximal speed for 30 minutes, the supernatant (nuclear fraction) was recovered. Cytoplasmic and nuclear fractions were measured for protein concentration by Bradford method and subjected to Western blot analysis.
Immunoprecipitation and western blotting
Experimental protocols for immunoprecipitation (IP) and immunoblotting follow those described previously.Citation49 Cells were lysed with RIPA lysis buffer (Cell Signaling Technology). Immunoprecipitation was performed with 2 ug of antibody against survivin, pan-ACK, AC-survivin (K129) or normal IgG (N IgG; as a negative control) in 1.0-mg whole-cell lysate. Cell lysates and/or immunoprecipitation cellular proteins were separated by SDS-PAGE in a 10% acrylamide gel and transferred onto nitrocellulose membrane for immunoblot.
Statistical analysis
SPSS (Statistic Package for Social Sciences) 13.0 for Windows (SPSS Inc., Chicago, IL, USA) was used to analyze data. The results are expressed as mean ± SEM. Statistical analysis was performed using the nonparametric Comparisons Test and Student's t-test. When the P-value was < 0.05, the difference was regarded as statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the doctoral Innovation Fund of Shanghai Jiao Tong University School of Medicine BXJ201625 (to SLL), by Shanghai Summit & Plateau Disciplines, by grant of National Nature Science Foundation of China 81572759 (to JD), 81620108022 (to JD), 81472516(to JZH), 81602367(to XY), grant of Ministry of Science and Technology No. 2013CB910900 (to JD), and by grants of Shanghai Municipal Planning Commission Clinical Center Project.
References
- Kademani D. Oral cancer. Mayo Clin Proc 2007; 82:878-87; PMID:17605971; http://dx.doi.org/10.4065/82.7.878
- Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol 2009; 45:309-16; PMID:18804401; http://dx.doi.org/10.1016/j.oraloncology.2008.06.002
- Schliephake H. Prognostic relevance of molecular markers of oral cancer–a review. Int J Oral Maxillofac Surg 2003; 32:233-45; PMID:12767868; http://dx.doi.org/10.1054/ijom.2002.0383
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 2007; 26:1324-37; PMID:17322918; http://dx.doi.org/10.1038/sj.onc.1210220
- Uren AG, Pakusch M, Hawkins CJ, Puls KL, Vaux DL. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc Natl Acad Sci U S A 1996; 93:4974-8; PMID:8643514; http://dx.doi.org/10.1073/pnas.93.10.4974
- Chiou SK, Jones MK, Tarnawski AS. Survivin - an anti-apoptosis protein: its biological roles and implications for cancer and beyond. Med Sci Monit 2003; 9:Pi25-9; PMID:12709681
- Blanc-Brude OP. Mesri M, Wall NR, Plescia J, Dohi T, Altieri DC. Therapeutic targeting of the survivin pathway in cancer: initiation of mitochondrial apoptosis and suppression of tumor-associated angiogenesis. Clin Cancer Res 2003; 9:2683-92; PMID:12855648
- Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC, Hwang JI, Chung CW, Jung YK, Oh BH. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry 2001; 40:1117-23; PMID:11170436; http://dx.doi.org/10.1021/bi001603q
- Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene 2003; 22:8581-9; PMID:14634620; http://dx.doi.org/10.1038/sj.onc.1207113
- O'Connor DS, Schechner JS, Adida C, Mesri M, Rothermel AL, Li F, Nath AK, Pober JS, Altieri DC. Control of apoptosis during angiogenesis by survivin expression in endothelial cells. Am J Patho 2000; 156:393-8; http://dx.doi.org/10.1016/S0002-9440(10)64742-6
- Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer 2003; 3:46-5; PMID:12509766; http://dx.doi.org/10.1038/nrc968
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997; 3:917-21; PMID:9256286; http://dx.doi.org/10.1038/nm0897-917
- Schimmer AD. Inhibitor of apoptosis proteins: translating basic knowledge into clinical practice. Cancer Res 2004; 64:7183-90; PMID:15492230; http://dx.doi.org/10.1158/0008-5472.CAN-04-1918
- Li F, Yang J, Ramnath N, Javle MM, Tan D. Nuclear or cytoplasmic expression of survivin: what is the significance? Int J Cancer 2005; 114:509-12; PMID:15578717; http://dx.doi.org/10.1002/ijc.20768
- Ling X, Yang J, Tan D, Ramnath N, Younis T, Bundy BN, Slocum HK, Yang L, Zhou M, Li F. Differential expression of survivin-2B and survivin-DeltaEx3 is inversely associated with disease relapse and patient survival in non-small-cell lung cancer (NSCLC). Lung Cancer 2005; 49:353-61; PMID:15936846; http://dx.doi.org/10.1016/j.lungcan.2005.03.037
- Lu B, Gonzalez A, Massion PP, Shyr Y, Shaktour B, Carbone DP, Hallahan DE. Nuclear survivin as a biomarker for non-small-cell lung cancer. Br J Cancer 2004; 91:537-40; PMID:15266313; http://dx.doi.org/10.1038/sj.bjc.6602027
- Shinohara ET, Gonzalez A, Massion PP, Chen H, Li M, Freyer AS, Olson SJ, Andersen JJ, Shyr Y, Carbone DP, et al. Nuclear survivin predicts recurrence and poor survival in patients with resected nonsmall cell lung carcinoma. Cancer 2005; 103:1685-92; PMID:15742356; http://dx.doi.org/10.1002/cncr.20951
- Vischioni B, van der Valk P, Span SW, Kruyt FA, Rodriguez JA, Giaccone G. Nuclear localization of survivin is a positive prognostic factor for survival in advanced non-small-cell lung cancer. Ann Oncol 2004; 15:1654-60; PMID:15520067; http://dx.doi.org/10.1093/annonc/mdh436
- Peng CH, Liao CT, Peng SC, Chen YJ, Cheng AJ, Juang JL, Tsai CY, Chen TC, Chuang YJ, Tang CY, et al. A novel molecular signature identified by systems genetics approach predicts prognosis in oral squamous cell carcinoma. PloS One 2011; 6:e23452; PMID:21853135; http://dx.doi.org/10.1371/journal.pone.0023452
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401-4; PMID:22588877; http://dx.doi.org/10.1158/2159-8290.CD-12-0095
- Wang H, Holloway MP, Ma L, Cooper ZA, Riolo M, Samkari A, Elenitoba-Johnson KS, Chin YE, Altura RA. Acetylation directs survivin nuclear localization to repress STAT3 oncogenic activity. J Biol Chem 2010; 285:36129-37; PMID:20826784; http://dx.doi.org/10.1074/jbc.M110.152777
- Fukuda S, Pelus LM. Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(+) cells by hematopoietic growth factors: implication of survivin expression in normal hematopoiesis. Blood 2001; 98:2091-100; PMID:11567995; http://dx.doi.org/10.1182/blood.V98.7.2091
- Chiou SK, Moon WS, Jones MK, Tarnawski AS. Survivin expression in the stomach: implications for mucosal integrity and protection. Biochem Biophys Res Commun 2003; 305:374-9; PMID:12745085; http://dx.doi.org/10.1016/S0006-291X(03)00724-1
- Adida C, Haioun C, Gaulard P, Lepage E, Morel P, Briere J, Dombret H, Reyes F, Diebold J, Gisselbrecht C, et al. Prognostic significance of survivin expression in diffuse large B-cell lymphomas. Blood 2000; 96:1921-5; PMID:10961895
- Azuhata T, Scott D, Takamizawa S, Wen J, Davidoff A, Fukuzawa M, Sandler A. The inhibitor of apoptosis protein survivin is associated with high-risk behavior of neuroblastoma. J Pediatr Surg 2001; 36:1785-91; PMID:11733907; http://dx.doi.org/10.1053/jpsu.2001.28839
- Ikeguchi M, Liu J, Kaibara N. Expression of survivin mRNA and protein in gastric cancer cell line (MKN-45) during cisplatin treatment. Apoptosis 2002; 7:23-9; PMID:11773702; http://dx.doi.org/10.1023/A:1013556727182
- Kim YH, Kim SM, Kim YK, Hong SP, Kim MJ, Myoung H. Evaluation of survivin as a prognostic marker in oral squamous cell carcinoma. J Oral Pathol Med 2010; 39:368-75; PMID:20050981
- Iyer SG, Pradhan SA, Pai PS, Patil S. Surgical treatment outcomes of localized squamous carcinoma of buccal mucosa. Head Neck 2004; 26:897-902; PMID:15390193; http://dx.doi.org/10.1002/hed.20096
- Rao DN, Shroff PD, Chattopadhyay G, Dinshaw KA. Survival analysis of 5595 head and neck cancers–results of conventional treatment in a high-risk population. Br J Cancer 1998; 77:1514-8; PMID:9652771; http://dx.doi.org/10.1038/bjc.1998.249
- Kennedy SM, O'Driscoll L, Purcell R, Fitz-Simons N, McDermott EW, Hill AD, O'Higgins NJ, Parkinson M, Linehan R, Clynes M. Prognostic importance of survivin in breast cancer. Br J Cancer 2003; 88:1077-83; PMID:12671708; http://dx.doi.org/10.1038/sj.bjc.6600776
- Lehner R, Lucia MS, Jarboe EA, Orlicky D, Shroyer AL, McGregor JA, Shroyer KR. Immunohistochemical localization of the IAP protein survivin in bladder mucosa and transitional cell carcinoma. Appl Immunohistochem Mol Morphol 2002; 10:134-8; PMID:12051631
- Okada E, Murai Y, Matsui K, Isizawa S, Cheng C, Masuda M, Takano Y. Survivin expression in tumor cell nuclei is predictive of a favorable prognosis in gastric cancer patients. Cancer Lett 2001; 163:109-16; PMID:11163114; http://dx.doi.org/10.1016/S0304-3835(00)00677-7
- Trieb K, Lehner R, Stulnig T, Sulzbacher I, Shroyer KR. Survivin expression in human osteosarcoma is a marker for survival. Eur J Surg Oncol 2003; 29:379-82; PMID:12711293; http://dx.doi.org/10.1053/ejso.2002.1415
- Grabowski P, Kühnel T, Mühr-Wilkenshoff F, Heine B, Stein H, Höpfner M, Germer CT, Scherübl H. Prognostic value of nuclear survivin expression in oesophageal squamous cell carcinoma. Br J Cancer 2003; 88:115-9; PMID:12556969; http://dx.doi.org/10.1038/sj.bjc.6600696
- Fortugno P, Wall NR, Giodini A, O'Connor DS, Plescia J, Padgett KM, Tognin S, Marchisio PC, Altieri DC. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci 2002; 115:575-85; PMID:11861764
- Mahotka C, Liebmann J, Wenzel M, Suschek CV, Schmitt M, Gabbert HE, Gabbert HE, Gerharz CD. Differential subcellular localization of functionally divergent survivin splice variants. Cell Death Differ 2002; 9:1334-42; PMID:12478470; http://dx.doi.org/10.1038/sj.cdd.4401091
- De Maria S, Pannone G, Bufo P, Santoro A, Serpico R, Metafora S, Rubini C, Pasquali D, Papagerakis SM, Staibano S, et al. Survivin gene-expression and splicing isoforms in oral squamous cell carcinoma. J Cancer Res Clin Oncol 2009; 135:107-16; PMID:18642030; http://dx.doi.org/10.1007/s00432-008-0433-z
- Barrett RM, Colnaghi R, Wheatley SP. Threonine 48 in the BIR domain of survivin is critical to its mitotic and anti-apoptotic activities and can be phosphorylated by CK2 in vitro. Cell Cycle 2011; 10:538-48; PMID:21252625; http://dx.doi.org/10.4161/cc.10.3.14758
- Colnaghi R, Wheatley SP. Liaisons between survivin and Plk1 during cell division and cell death. J Biol Chem 2010; 285:22592-604; PMID:20427271; http://dx.doi.org/10.1074/jbc.M109.065003
- Nousiainen M, Silljé HH, Sauer G, Nigg EA, Körner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A 2006; 103:5391-6; PMID:16565220; http://dx.doi.org/10.1073/pnas.0507066103
- Wheatley SP, Barrett RM, Andrews PD, Medema RH, Morley SJ, Swedlow JR, Lens SM. Phosphorylation by aurora-B negatively regulates survivin function during mitosis. Cell Cycle 2007; 6:1220-30; PMID:17457057; http://dx.doi.org/10.4161/cc.6.10.4179
- O'Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci U S A 2000; 97:13103-7; PMID:11069302; http://dx.doi.org/10.1073/pnas.240390697
- Nogueira-Ferreira R, Vitorino R, Ferreira-Pinto MJ, Ferreira R, Henriques-Coelho T. Exploring the role of post-translational modifications on protein-protein interactions with survivin. Arch Biochem Biophys 2013; 538:64-70; PMID:23938875; http://dx.doi.org/10.1016/j.abb.2013.07.027
- Zhu Y, Gu YX, Mo JJ, Shi JY, Qiao SC, Lai HC. N-acetyl cysteine protects human oral keratinocytes from Bis-GMA-induced apoptosis and cell cycle arrest by inhibiting reactive oxygen species-mediated mitochondrial dysfunction and the PI3K/Akt pathway. Toxicol In Vitro 2015; 29:2089-101; PMID:26343756; http://dx.doi.org/10.1016/j.tiv.2015.09.002
- Liu SL, Zhong SS, Ye DX, Chen WT, Zhang ZY, Deng J. Repression of G protein-coupled receptor family C group 5 member A is associated with pathologic differentiation grade of oral squamous cell carcinoma. J Oral Pathol Med 2013; 42:761-8; PMID:23651229; http://dx.doi.org/10.1111/jop.12077
- Liu S, Ye D, Guo W, Yu W, He Y, Hu J, Wang Y, Zhang L, Liao Y, Song H, et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget 2015; 6:6887-901; PMID:25749385; http://dx.doi.org/10.18632/oncotarget.3159
- Liu S, Liu L, Ye W, Ye D, Wang T, Guo W, Liao Y, Xu D, Song H, Zhang L, et al. High Vimentin Expression Associated with Lymph Node Metastasis and Predicated a Poor Prognosis in Oral Squamous Cell Carcinoma. Sci Rep 2016; 6:38834; PMID:27966589; http://dx.doi.org/10.1038/srep38834
- Li J, Zhou BP. Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 2011; 11:49; PMID:21284870; http://dx.doi.org/10.1186/1471-2407-11-49
- Zhong S, Yin H, Liao Y, Yao F, Li Q, Zhang J, Jiao H, Zhao Y, Xu D, Liu S, et al. Lung Tumor Suppressor GPRC5A Binds EGFR and Restrains Its Effector Signaling. Cancer Res 2015; 75:1801-14; PMID:25744720; http://dx.doi.org/10.1158/0008-5472.CAN-14-2005
