Cells must tightly coordinate the levels of many of their proteins to navigate accurately and safely the transitions of the cell cycle. For cells to grow and divide, not only proteins but also lipids must be synthesized anew in every cell cycle. The amount, composition, and localization of the lipid repertoire are dynamic in dividing cells.Citation1 However, little is known about how cells regulate their lipid content during a cell cycle. Here we highlight results from genome-wide studies in yeastCitation2 and humanCitation3,4 cells that revealed surprising mechanisms of control, at the translational level, of lipid metabolism in the cell cycle.
To find transcripts under translational control, ribosome profiling quantifies by deep sequencing all pieces of mRNAs in the cell bound to translating ribosomes.Citation5 This technique has now been used to find cases of gene-specific, cell cycle-dependent, translational control.Citation2-4 Different studies used different systems and methodologies to obtain synchronous samples. Blank and colleagues used budding yeast collected at specific sizes via centrifugal elutriation to examine by ribosome profiling a cell size series spanning the entire cell cycle.Citation2 The synchrony achieved is free of possible arrest-induced artifacts and preserves as much as possible the normal coordination of growth and division. A striking result was that translation of mRNAs encoding the core enzymes of lipid biogenesis, acetyl-CoA carboxylase (ACC1) and fatty acid synthase (FAS1 and FAS2), was upregulated in mitosis.Citation2 A short upstream open reading frame (uORF) adjusts the translation of ACC1, leading to >10-fold increase late in the cell cycle, and also represses translation of ACC1 in poor media.Citation2
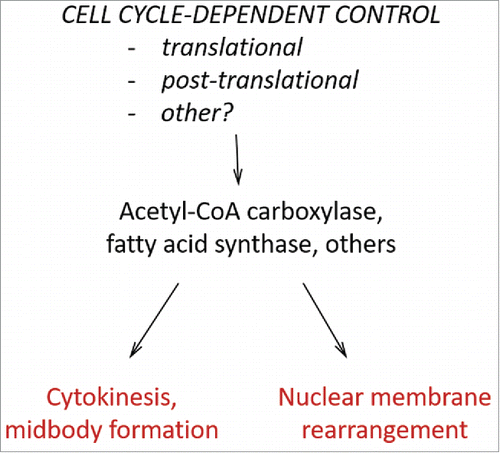
Human cells arrested at different points along the cell cycle were also subjected to ribosome profiling.Citation3,4 The cells in the studies of Stumpf and colleaguesCitation3 were not released from their arrest, leaving open the possibility of artifacts. Nonetheless, translation of mRNAs encoding enzymes of lipid metabolism was regulated, with most of them peaking in mitosis.Citation3 The human cells examined by Tanenbaum and colleagues were arrested in the G2 phase with a small-molecule inhibitor of the cyclin-dependent kinase CDK1.Citation4 Washing the inhibitor away enabled the arrested cells to progress synchronously through mitosis, and enter the next G1. It is not clear if the cells in this experiment attained their normal degree of coupling between growth and division since the arrest period was 18 h and the cells were released for only 45 or 225 min.Citation4 Despite these limitations, Tanenbaum and colleagues interrogated progress through a key cell cycle phase, mitosis, during which animal cells repress overall protein synthesis.Citation4 Most (> 90%) of the mRNAs they identified were repressed translationally in mitosis.Citation4 Under the same conditions, demonstrating the varying nature of transcript-specific translational control, some mRNAs had increased translational efficiency.Citation4 Among them was FASN, encoding human fatty acid synthase, whose translational efficiency was increased by >2-fold in mitosis compared with the G2 phase.Citation4 But mitotic upregulation of lipid metabolism need not come about only through translational control. De novo fatty acid synthesis and upregulation of human acetyl-CoA carboxylase (ACACA) through post-translational control were essential for completion of mitosis ().Citation6
There could be many reasons why cells need new lipids late in the cell cycle. The most obvious need would arise from the sudden increase in the outer cell surface upon exit from mitosis, which approaches ≈40% for spherical cells. We note, however, that yeast cells with perturbed lipid homeostasis were still able to increase their exterior surface during mitosis,Citation2 arguing for more specialized roles for lipids in the eukaryotic cell cycle. A comprehensive lipidomic study by Atilla-Gokcumen and colleagues demonstrated extensive changes in lipid composition and localization during the cell cycle in human cells, especially along the midbody before cell separation.Citation1 They also found 23 lipid biosynthetic enzymes to be essential for cytokinesis, including enzymes of sphingolipid metabolism and fatty acid elongases.Citation1 Finally, the nuclear membrane also goes through dramatic rearrangements during cell division, from complete breakdown and re-assembly in animal cells, to massive expansion during the closed mitosis that many fungi undergo. De novo lipid biogenesis is needed for the expansion of the nuclear membrane in yeast. Reduced function of Polo-like kinase, acetyl-CoA carboxylase or fatty acid synthase was proposed to lower phosphatidic acid levels, reducing the ability of cells to increase the area of their nuclear membrane.Citation7
Overall, the studies we highlighted point to the emerging role of lipid metabolism in underpinning cell cycle landmarks and the ability of cells to progress through cell cycle transitions. Interest in the area is high, extending to therapeutic applications, with inhibitors of fatty acid synthase in Phase 1 clinical trials for the management of advanced stage solid tumors (ClinicalTrials.gov Identifier: NCT02223247). Translational control expands the ways that cells control their lipid composition and it may link 2 fundamental aspects of cell growth in volume and surface, synthesis of proteins and lipids. Future work will connect specific lipids and the enzymes that make them with their corresponding structural or signaling roles in the cell cycle.
Figure 1. Cell cycle-dependent control of lipid metabolism drives cell cycle transitions and formation of cell cycle landmarks. Expression of enzymes of lipid metabolism, such as acetyl-CoA carboxylase and fatty acid synthase, is regulated in the cell cycle by several mechanisms (including translational control), underscoring lipid requirements for progress through cell cycle transitions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Atilla-Gokcumen GE, Muro E, Relat-Goberna J, Sasse S, Bedigian A, Coughlin ML, Garcia-Manyes S, Eggert US. Dividing cells regulate their lipid composition and localization. Cell 2014; 156:428-39; PMID:24462247; http://dx.doi.org/10.1016/j.cell.2013.12.015
- Blank HM, Perez R, He C, Maitra N, Metz R, Hill J, Lin Y, Johnson CD, Bankaitis VA, Kennedy BK, et al. Translational control of lipogenic enzymes in the cell cycle of synchronous, growing yeast cells. EMBO J 2017; 36:487-502; PMID:28057705; http://dx.doi.org/10.15252/embj.201695050
- Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. The translational landscape of the mammalian cell cycle. Mol Cell 2013; 52:574-82; PMID:24120665; http://dx.doi.org/10.1016/j.molcel.2013.09.018
- Tanenbaum ME, Stern-Ginossar N, Weissman JS, Vale RD. Regulation of mRNA translation during mitosis. Elife 2015; 4:e07957; http://dx.doi.org/10.7554/eLife.07957
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 2012; 7:1534-50; PMID:22836135; http://dx.doi.org/10.1038/nprot.2012.086
- Scaglia N, Tyekucheva S, Zadra G, Photopoulos C, Loda M. De novo fatty acid synthesis at the mitotic exit is required to complete cellular division. Cell Cycle 2014; 13:859-68; PMID:24418822; http://dx.doi.org/10.4161/cc.27767
- Walters AD, May CK, Dauster ES, Cinquin BP, Smith EA, Robellet X, D'Amours D, Larabell CA, Cohen-Fix O. The yeast polo kinase Cdc5 regulates the shape of the mitotic nucleus. Current Biology : CB 2014; 24:2861-7; PMID:25454593; http://dx.doi.org/10.1016/j.cub.2014.10.029
