ABSTRACT
Cooperation between cellular organelles such as mitochondria, peroxisomes and the ER is essential for a variety of important and diverse metabolic processes. Effective communication and metabolite exchange requires physical linkages between the organelles, predominantly in the form of organelle contact sites. At such contact sites organelle membranes are brought into close proximity by the action of molecular tethers, which often consist of specific protein pairs anchored in the membrane of the opposing organelles. Currently numerous tethering components have been identified which link the ER with multiple other organelles but knowledge of the factors linking the ER with peroxisomes is limited. Peroxisome-ER interplay is important because it is required for the biosynthesis of unsaturated fatty acids, ether-phospholipids and sterols with defects in these functions leading to severe diseases. Here, we characterize acyl-CoA binding domain protein 4 (ACBD4) as a tail-anchored peroxisomal membrane protein which interacts with the ER protein, vesicle-associated membrane protein-associated protein–B (VAPB) to promote peroxisome-ER associations.
Introduction
The study of organelle interactions at membrane contact sites is an area of cell biology which has expanded rapidly over the last decade due to the understanding that interorganellar communication is vital for cellular function. A striking example of the importance of organelle interplay is found in the relationship between the endoplasmic reticulum (ER) and peroxisomes (reviewed in ref. Citation1). These 2 organelles have been known to be intimately associated since ultrastructural studies in the 1960s detected close apposition between ER tubules and peroxisomal membranes.Citation2,3 A number of metabolic pathways require the combined action of both peroxisomal and ER-resident enzymes. Most notably in the production of ether-phospholipids such as plasmalogens which requires generation of a characteristic ether bond by peroxisomal enzymes before the remaining steps in biosynthesis can be completed in the ER.Citation4,Citation5 Failure to properly assemble peroxisomes (e.g. in Zellweger spectrum disorders),Citation6 mutations in the genes which encode the peroxisomal enzymes or import factors which bring the enzymes into peroxisomes result in a deficiency in ether phospholipid production and lead to diseases such as rhizomelic chondrodysplasia punctata (RCDP).Citation7-9 In mammals, as well as linking with peroxisomes for metabolic cooperation, the ER can also play a role, perhaps in collaboration with mitochondria, in the de novo generation of peroxisomes.Citation10-12 The full extent the ER plays in peroxisome biogenesis is unclear but appears to at least involve the provision of membrane phospholipids (and potentially membrane proteins such as Pex16) for formation of the peroxisomal membrane.Citation13,Citation14
As well as interacting with peroxisomes, the ER forms contact sites with mitochondria, Golgi complex, plasma membrane, and endosomesCitation15-17 (see ref. Citation18 for a comprehensive list). Two key players are vesicle-associated membrane protein-associated proteins – A and B (VAPA/B), which are present in several important contact sites involving the ER.Citation19 VAPA/B are ER-resident membrane proteins containing a major sperm protein (MSP) domain that interacts with proteins containing a FFAT or FFAT-like motif.Citation20 One such protein is PTPIP51, a mitochondrial membrane protein which interacts with VAPB to mediate mitochondria-ER associations, facilitating calcium exchange and regulating autophagy.Citation17,Citation21
Recently, we identified peroxisomal acyl-CoA binding domain protein 5 (ACBD5) and VAPB as interaction partners of a molecular tether which physically links peroxisomes to the ER in mammals.Citation22 Both VAPB and ACBD5 are C-tail-anchored (TA) membrane proteins, defined as proteins which contain N-terminal functional domains followed by a single transmembrane domain (TMD) close to the C-terminus and a short C-terminal tail region. These characteristic properties dictate that TA proteins are post-translationally sorted to their target membrane with the N-terminus facing the cytosol.Citation23 In another recent study we investigated the targeting properties of TA proteins, discovering the importance of interplay between TMD hydrophobicity and tail-charge, and developed a statistical model to predict cellular localization of TA proteins based on these physicochemical parameters.Citation24 Using this bioinformatics prediction tool we identified an isoform of ACBD4 (isoform 2), a predicted TA protein of unknown function and localization, as a potential peroxisomal protein and confirmed this by expression of Myc-ACBD4iso2 in COS-7 cells.Citation24 ACBD4, like ACBD5, is a member of the ACBD family which is characterized by the presence of an acyl-CoA binding domain. Seven different ACBDs have been identified in mammals but the acyl-CoA binding protein structural fold has been found in 48 different protein architectures across all species.Citation25 Thus, although ACBD4 and ACBD5 share 58% sequence identity this is mainly isolated to similarities in the N-terminal acyl-CoA binding domain, with the rest of the proteins showing significant differences.
Here, we show that ACBD4 isoform2 is a tail-anchored peroxisomal protein which interacts with the ER-resident protein VAPB to facilitate interaction between the 2 organelles. These results suggest that ACBD4, like ACBD5, can act as a molecular tether, physically linking peroxisomes and the ER making this the second protein involved in peroxisome-ER contacts in mammals.
Results
ACBD4iso2 is a C-tail-anchored membrane protein which shows peroxisomal targeting when expressed in COS-7 cells. ACBD4 has 3 major isoforms (as defined by UniProt identifier: Q8NC06) one of which, isoform 2 (UniProt identifier: Q8NC06–2), is predicted to contain a C-terminal TMD and tail. In addition to the characteristic N-terminal acyl-CoA binding domain, other predicted structural features in ACBD4iso2 include a potential coiled-coil domain and a predicted FFAT-like motif (). Previously, we showed that Myc-ACBD4iso2 expressed in COS-7 cells localized to peroxisomes.Citation24 Here, we further characterize ACBD4 localization showing that while we always observe Myc-ACBD4iso2 targeting to peroxisomes (), when expression levels are high we observe changes in peroxisome morphology () and weak, non-peroxisomal signal () which co-localizes with a mitochondrial marker (). This phenomenon has also been observed for other peroxisomal TA proteins such as Pex26Citation26 which also shows mitochondrial localization when expression is high.
Figure 1 . ACBD4iso2 is a peroxisomal C-tail-anchored protein. (A) Schematic overview of ACBD4iso2 domain structure. ACBD = acyl-CoA binding domain, FFAT-like = 2 phenyalanines in an acidic tract, CC = coiled coil, TMD = transmembrane domain. (B-E) Subcellular localization patterns for ACBD4iso2. COS-7 cells transfected with Myc-ACBD4iso2 were immunolabelled using αPEX14 (peroxisomal marker), αTOM20 (mitochondrial marker) and αMyc antibodies. (E) Higher magnifications of boxed regions are shown (F-G) Differential permeabilisation. COS-7 cells expressing FLAG-ACBD4iso2 were fixed, permeabilised with either Triton X-100 (0.2% in PBS) (F) or digitonin (2.5µg/ml in PBS) (G), and stained with αCatalase (PO matrix), αPEX14 (PO membrane) or αFLAG antibodies. Bars, 10 µm (overlay), 2µm (magnified sections).
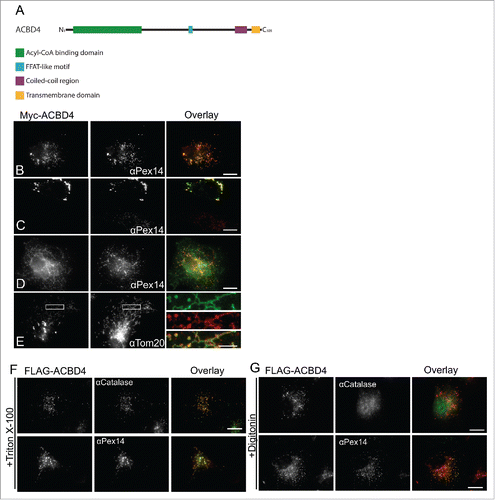
To confirm that ACBD4iso2 is a C-tail-anchored protein with the N-terminus exposed to the cytosol we performed differential permeabilisation experiments using either digitonin or Triton X-100. Triton X-100 permeabilises peroxisomal membranes whereas upon digitonin treatment peroxisome membranes remain intact.Citation27,Citation28 Accordingly, following digitonin treatment the peroxisomal matrix marker catalase was inaccessible to antibodies and was only detected after Triton X-100 treatment (). After digitonin treatment the N-terminal FLAG-tag of FLAG-ACBD4iso2 was detectable using FLAG antibodies indicating that the N-terminus of ACBD4 is exposed to the cytosol () similar to what was found for ACBD5.Citation24
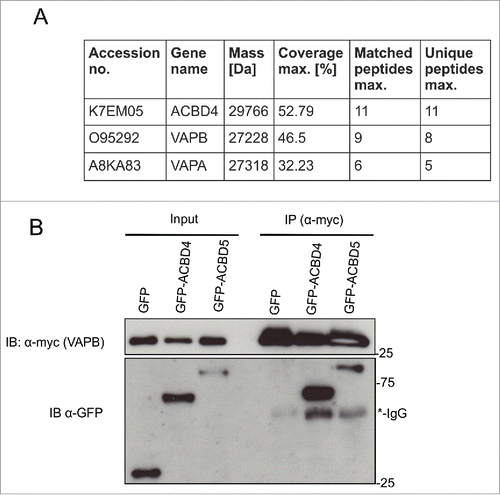
ACBD4iso2 interacts with VAPB. As the function of ACBD4 is unknown we performed proteomics studies to identify potential binding partners. GFP-ACBD4iso2 and GFP alone were expressed in COS-7 cells and pull down studies and mass spectrometry (MS) analyses were performed in triplicate. Following filtering of the results (based on a previous studyCitation29 only protein IDs with >1 unique peptide hits, >20% peptide:protein coverage and overall MS scores >30, which did not appear in any of the GFP only control experiments, were considered) from MS experiments we identified the ER membrane proteins VAPA and VAPB as candidate binding partners (). We next confirmed the ACBD4-VAPB interaction by immunoprecipitation (IP). GFP-ACBD4iso2 and Myc-VAPB were co-expressed in COS-7 cells and their interaction was assessed by IP using Myc-TRAP magnetic agarose beads (). As a positive control we used GFP-ACBD5 which we had previously shown to interact with Myc-VAPB using the same assay.Citation22 Using this assay we were able to confirm interaction between ACBD4iso2 and VAPB.
Figure 2 . ACBD4iso2 interacts with VAPB. (A) Identification of VAPB and VAPA by MS after co-immunoprecipitation (IP) with GFP-ACBD4iso2 from COS-7 cells (results from 3 experiments); GFP used as control. Only protein IDs which did not appear in any of the GFP only control experiments were considered. (B) Immunoprecipitation (IP) of GFP-ACBD4iso2 and Myc-VAPB after co-expression in COS-7 cells. GFP used as a negative control and GFP-ACBD5 as a positive control. Samples were immunoprecipitated (GFP-Trap) and immunoblotted (IB) using Myc/GFP antibodies.
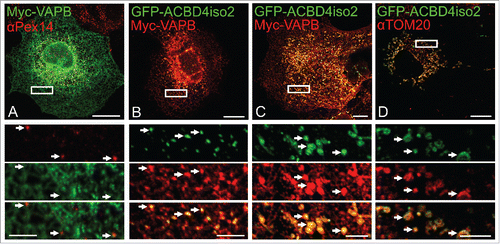
Co-expression of GFP-ACBD4iso2/Myc-VAPB promotes ER-PO associations. Having established that ACBD4 can interact with VAPB we wanted to test if ACBD4, like ACBD5, can play a role in mediating peroxisome-ER associations. To test this, we co-expressed Myc-VAPB and GFP-ACBD4iso2 in COS-7 cells and analyzed ER-PO localization using confocal microscopy (). In our previous study we observed that when both ACBD5 and VAPB were overexpressed we could observe increased ER-peroxisome associations which, strikingly, allowed vizualization of discrete peroxisomal structures when using VAPB as an ER marker.Citation22 Here, this characteristic PO-ER association was also observed when ACBD4 and VAPB were co-expressed together but not individually (). Furthermore, when we examined cells in which ACBD4 was found at mitochondria (see ) we detected increased association of VAPB-labeled ER with the mitochondrial marker, suggesting that in this case mis-targeted ACBD4 was mediating increased ER-mitochondria interactions (). These findings support a role for ACBD4 and VAPB interaction in ER-peroxisome tethering.
Figure 3 . ACBD4iso2/VAPB co-expression promotes PO-ER association. COS-7 cells were transfected with (A) Myc-VAPB alone (immunolabelled using αPEX14, a peroxisomal marker), (B) Myc-VAPB co-expressed with GFP-ACBD4iso2, (C) Myc-VAPB co-expressed with GFP-ACBD4iso2 showing mitochondrial mistargeting. (D) Co-localization of GFP-ACBD4iso2 with Tom20 (mitochondrial marker). Arrows highlight PO-ER association. Bars, 20 µm (overview), 5 µm (cut outs).
Discussion
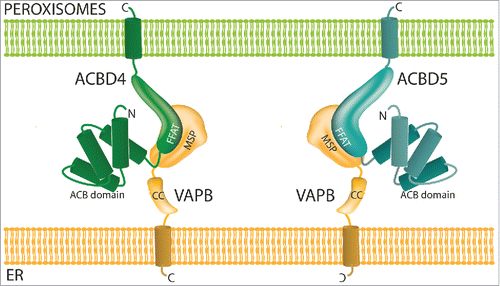
The data presented here, namely that ACBD4 is localized to peroxisomes and interacts with the ER protein VAPB to promote ER-peroxisome associations support the assumption that ACBD4 is acting as a tether.Citation30 In a recent publication we identified the first molecular mechanism which allowed peroxisome-ER interactions in mammalian cells via a tether consisting of peroxisomal ACBD5 and ER-resident VAPBCitation22 (). We showed that in the absence of ACBD5/VAPB peroxisomal membrane expansion was reduced, suggesting that the lipid flow from the ER to peroxisomes required for peroxisomal membrane growth was disrupted. In addition the movement of peroxisomes was increased. Simultaneously the group of Peter Kim used a parallel approach to reach the same conclusions, additionally reporting that plasmalogen synthesis is impaired when the VAPB-ACBD5 tether is disrupted.Citation31 Recent studies have now identified patients carrying pathogenic mutations which lead to the loss of ACBD5 protein.Citation32-34 In these cases increased levels of very-long-chain fatty acids (VLCFAs) were detected in patient cells likely due to reduced import into peroxisomes. This suggested a role for ACBD5 in binding VLCFAs in the cytosol and facilitating their transport into peroxisomes which would then be mediated by the peroxisomal ABC transporters at the peroxisomal membrane.Citation35,Citation36 It is not clear if ACBD5 interacts with the ABC transporters, with a recent study failing to identify ACBD5 as an interacting partner of ABCD2,Citation37 nor how the tethering function of ACBD5 is linked to its function in β-oxidation of VLCFAs. However, as ACBD4 also contains an acyl-CoA binding domain and a predicted FFAT-like motif it is tempting to speculate that ACBD4 may play a similar role to ACBD5 (). The differences between the 2 proteins may lie in substrate specificity, expression profile, regulation or type of tether. Yagita and colleaguesCitation34 reported that ACBD5 is able to preferentially bind VLCFAs in vitro but its optimal substrate was not identified and may differ from the optimal substrate for ACBD4. In our previous studyCitation22 knockdown of ACBD5 showed significant effects on the extent of peroxisome-ER interactions in HepG2 cells. As ACBD4 is reported to be expressed in these cells,Citation38 it is unlikely that normal ACBD4 levels can fully complement the function of ACBD5. It is possible that ACBD5 is the major tether for peroxisome-ER contacts whereas ACBD4 may play a role in a more specialized ER-peroxisome association. The presence of more than one tether which can link peroxisomes and the ER is in line with the multiple different tether combinations used by other organelles to cater for specialized functions.Citation39 Future studies will address these points and contribute to the understanding of the roles of ACBD4 and ACBD5 in peroxisome-ER interplay, lipid metabolism and how their dysfunction links to disease.
Figure 4 . Model of ACBD4/ACBD5-VAPB interaction. ACBD4 and ACBD5 are both C-tail anchored peroxisomal membrane proteins with functional domains in the cytoplasm which can interact with the MSP domain of ER-resident VAPB via a FFAT-like motif. ACB = acyl-CoA binding, FFAT = 2 phenyalanines in an acidic tract, MSP = major sperm protein binding domain.
Materials and Methods
Plasmids and antibodies
Myc-VAPB plasmid was kindly provided by C. Miller (King's College London, UK). A human ACBD4iso2 cDNA clone (Cusabio Life Sciences, http://www.cusabio.com/Clone/ACBD4–158327.html) was used as a template to generate GFP-ACBD4iso2 and FLAG-ACBD4iso2 using eGFP-C1 and pCMV-2B vectors respectively. Primers: ACBD4_iso2_GFP_For = AAACTCGAGCTATGGGCACCGAG AAAGAAAGCCCAGAGCCCGAC, ACBD4_iso2_GFP_Rev = TTGGATCCTCACCTC TTTTGGGTCCGAAACATTCGGAAGAGCC (XhoI, BamHI digest into eGFP-C1).
ACBD4_myc_For = AAGGATCCATGGGCACCGAGAAAGAAAGCCCAGAGCCCGAC, ACBD4iso2_myc_Rev = CTCTCGAGTCACCTCTTTTGGGTCCGAAACATTCGGAAGA GCC (XhoI, BamHI digest into pCMV2B). Antibodies were as follows: polyclonal rabbit anti-PEX14 (kindly provided by D. Crane, Griffith University, Brisbane, Australia); anti-catalase (Abcam, http://www.abcam.com/catalase-antibody-ab88650.html); anti-GFP (Thermofisher, https://www.thermofisher.com/antibody/product/GFP-Tag-Antibody-Polyclonal/A-11122); anti-Myc (Abcam, http://www.abcam.com/myc-tag-antibody-ab9106.html); anti-FLAG (SIGMA, http://www.sigmaaldrich.com/catalog/product/sigma/f3165?lang = enandregion = GB).
Cell culture and transfection
COS-7 cells (African green monkey kidney cells; ATCC, https://www.lgcstandards-atcc.org/products/All/CRL-1651) were cultured in DMEM, high glucose (4.5 g/L) supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C with 5% CO2 and 95% humidity. COS-7 cells were transfected using diethylaminoethyl (DEAE)-dextran (Sigma-Aldrich, http://www.sigmaaldrich.com/catalog/product/sigma/d9885) as described.Citation40
Immunofluorescence and microscopy
Cells were processed for immunofluorescence 24 h after transfection as described previously.Citation41 Cell imaging was performed using an Olympus IX81 microscope equipped with an UPlanSApo 100x/1.40 Oil objective (Olympus Optical, Hamburg, Germany), eGFP ET filter-set (470/40 ET Bandpass filter, Beamsplitter T495 LPXR and 525/50 ET Bandpass filter (Chroma Technology GmbH, Olching, Germany)), and TxRed HC Filter Set (562/40 BrightLine HC Beamsplitter HC BS 593, 624/40 BrightLine HC (Semrock, Rochester, USA)). Digital images were taken with a CoolSNAP HQ2 CCD camera and adjusted for contrast and brightness using MetaMorph 7 (Molecular Devices, https://www.moleculardevices.com/systems/metamorph-research-imaging/metamorph-microscopy-automation-and-image-analysis-software). Confocal images were obtained using a Leica SP8 equipped with: Argon laser (488), DPSS561 laser (561), HC PL APO 63x/1.3 Oil objective, HC PL APO 100x/1.44 Oil objective, Hybrid detectors (HyD).
Immunoprecipitation
GFP-ACBD4iso2, or GFP only control, and Myc-VAPB were expressed in COS-7 cells. After 48 h cells were washed in PBS and lysed in ice-cold lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1% NP-40, 1 mM PMSF and mini protease inhibitor cocktail (Roche, http://www.sigmaaldrich.com/catalog/product/roche/11836170001)). Unsolubilised material was pelleted by centrifugation at 100,000 × gav. Clarified lysates were then mixed with Myc-TRAP magnetic agarose beads (ChromoTek, http://www.chromotek.com/products/nano-traps/myc-trapr/) and incubated for 2 h at 4°C. Beads were washed extensively with lysis buffer and bound proteins were either eluted with Laemmli buffer or further processed for mass spectrometry analysis. Immunoprecipitates and total lysates were analyzed by Western immunoblotting.
Mass spectrometry (MS)
For MS analysis, immunoprecipitations (see above) from 3 independent experiments were analyzed for both GFP-ACBD4iso2 and a GFP only control. Sample preparation and protein identification were performed by the University of Bristol Proteomics Facility as described previously.Citation29 Extracted MS/MS spectra were searched against the Uniprot Human database and were filtered at 5% FDR. Additional filtering parameters were based on a previous study.Citation29 Only protein IDs with >1 unique peptide hits, >20% peptide:protein coverage and overall MS scores >30, which did not appear in any of the GFP only control experiments, were considered.
Abbreviations
| ACBD | = | acyl-CoA binding domain |
| IP | = | immunoprecipitation |
| PO | = | peroxisomes |
| VAPB | = | vesicle-associated membrane protein-associated protein–B |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
Author contributions
TS performed experiments; MS, MI, JC, and IC conceived the project, performed experiments, and analyzed data; JC, MS and MI wrote the manuscript.
Acknowledgments
We thank all colleagues who provided plasmids and antibodies, and T Levine for sharing data.
Funding
This work was supported by BBSRC (BB/K006231/1, BB/N01541X/1). MS is supported by the Marie Curie Initial Training Network action PerFuMe (316723). The authors declare no competing financial interests.
References
- Schrader M, Godinho LF, Costello JL, Islinger M. The different facets of organelle interplay—an overview of organelle interactions. Front Cell Dev Biol 2015; 3:56; PMID:26442263; https://doi.org/10.3389/fcell.2015.00056
- Novikoff A, Shin WY. The endoplasmic reticulum in the Golgi zone and its relation to microbodies, Golgi apparatus and autophagic vacuoles in rat liver cells. J Microsc 1964; 3:187–206
- Yamamoto K, Fahimi HD. Three-dimensional reconstruction of a peroxisomal reticulum in regenerating rat liver: evidence of interconnections between heterogeneous segments. J Cell Biol 1987; 105:713–22; PMID:2887576; https://doi.org/10.1083/jcb.105.2.713
- Dorninger F, Brodde A, Braverman NE, Moser AB, Just WW, Forss-Petter S, Brügger B, Berger J. Homeostasis of phospholipids - The level of phosphatidylethanolamine tightly adapts to changes in ethanolamine plasmalogens. Biochim Biophys Acta 2015; 1851:117–128; PMID:25463479; https://doi.org/10.1016/j.bbalip.2014.11.005
- Wanders RJA, Waterham HR, Ferdinandusse S. Metabolic interplay between Peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. Front Cell Dev Biol Front Cell Dev Biol 2016; 3:833383–9
- Braverman NE, Raymond GV, Rizzo WB, Moser AB, Wilkinson ME, Stone EM, Steinberg SJ, Wangler MF, Rush ET, Hacia JG, et al. Peroxisome biogenesis disorders in the Zellweger spectrum: An overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab 2016; 117:313–21; PMID:26750748; https://doi.org/10.1016/j.ymgme.2015.12.009
- Braverman N, Steel G, Obie C, Moser A, Moser H, Gould SJ, Valle D. Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat Genet 1997; 15:369–76; PMID:9090381; https://doi.org/10.1038/ng0497-369
- Heikoop JC, Van Roermund CWT, Just WW, Ofman R, Schutgens RBH, Heymans HSA, Wanders RJA, Tager JM. Rhizomelic chondrodysplasia punctata. Deficiency of 3-oxoacyl-coenzyme A thiolase in peroxisomes and impaired processing of the enzyme. J Clin Invest 1990; 86:126–30; PMID:2365812; https://doi.org/10.1172/JCI114674
- Motley AM, Brites P, Gerez L, Hogenhout E, Haasjes J, Benne R, Tabak HF, Wanders RJA, Waterham HR. Mutational spectrum in the PEX7 gene and functional analysis of mutant alleles in 78 patients with rhizomelic chondrodysplasia punctata type 1. Am J Hum Genet 2002; 70:612–24; PMID:11781871; https://doi.org/10.1086/338998
- Hettema EH, Erdmann R, van der Klei IJ, Veenhuis M. Evolving models for peroxisome biogenesis. Curr Opin Cell Biol 2014; 29:25–30; PMID:24681485; https://doi.org/10.1016/j.ceb.2014.02.002
- Rucktäschel R, Halbach A, Girzalsky W, Rottensteiner H, Erdmann R. De novo synthesis of peroxisomes upon mitochondrial targeting of Pex3p. Eur J Cell Biol 2010; 89:947–54; PMID:20655617; https://doi.org/10.1016/j.ejcb.2010.06.012
- Sugiura A, Mattie S, Prudent J, McBride HM. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 2017; 542:251–4; PMID:28146471; https://doi.org/10.1038/nature21375
- Aranovich A, Hua R, Rutenberg AD, Kim PK. PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J Cell Sci 2014; 127:3675–86; PMID:25002403; https://doi.org/10.1242/jcs.146282
- Raychaudhuri S, Prinz WA. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc Natl Acad Sci USA 2008; 105:15785–90; PMID:18836080; https://doi.org/10.1073/pnas.0808321105
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio MC, et al. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J Cell Sci 2013; 126:5500–12; PMID:24105263; https://doi.org/10.1242/jcs.139295
- Doghman-Bouguerra M, Lalli E. ER-mitochondria contacts find their FATE. Cell Cycle 2016; 15, 3159–3160; PMID:27579833; https://doi.org/10.1080/15384101.2016.1220720
- Stoica R, De Vos KJ, Paillusson S, Mueller S, Sancho RM, Lau KF, Vizcay-Barrena G, Lin WL, Xu YF, Lewis J, et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun 2014; 5:3996; PMID:24893131
- Eisenberg-Bord M, Shai N, Schuldiner M, Bohnert M. A Tether Is a Tether Is a Tether: Tethering at membrane contact sites. Dev Cell 2016; 39:395–409; PMID:27875684; https://doi.org/10.1016/j.devcel.2016.10.022
- Murphy SE, Levine TP. VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta Mol Cell Biol Lipids 2016; 8:952–61; https://doi.org/10.1016/j.bbalip.2016.02.009
- Loewen CJR, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J 2003; 22:2025–35; PMID:12727870; https://doi.org/10.1093/emboj/cdg201
- Gomez-Suaga P, Paillusson S, Stoica R, Noble W, Hanger DP, Miller CCJ. The ER-mitochondria tethering complex VAPB- PTPIP51 regulates autophagy. Curr Biol 2017; 27:371–85; PMID:28132811; https://doi.org/10.1016/j.cub.2016.12.038
- Costello JL, Castro IG, Hacker C, Schrader TA, Metz J, Zeuschner D, Azadi AS, Godinho LF, Costina V, Findeisen P, et al. ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J. Cell Biol 2017a; 216:331–42; https://doi.org/10.1083/jcb.201607055
- Borgese N, Fasana E. Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta 2011; 1808:937–46; PMID:20646998; https://doi.org/10.1016/j.bbamem.2010.07.010
- Costello JL, Castro IG, Camoes F, Schrader TA, McNeall D, Yang J, Giannopoulou EA, Gomes S, Pogenberg V, Bonekamp NA, et al. Predicting the targeting of tail-anchored proteins to subcellular compartments in mammalian cells. J Cell Sci 2017; pii: jcs.200204. [Epub ahead of print]; PMID: 28325759; https://doi.org/10.1242/jcs.200204
- Neess D, Bek S, Engelsby H, Gallego SF, Færgeman NJ. Long-chain acyl-CoA esters in metabolism and signaling: Role of acyl-CoA binding proteins. Prog Lipid Res 2015; 59:1–25; PMID:25898985; https://doi.org/10.1016/j.plipres.2015.04.001
- Halbach A, Landgraf C, Lorenzen S, Rosenkranz K, Volkmer-Engert R, Erdmann R, Rottensteiner H. Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J Cell Sci 2006; 119:2508–17; PMID:16763195; https://doi.org/10.1242/jcs.02979
- Schrader M, Reuber BE, Morrell JC, Jimenez-sanchez G, Obie C, Stroh TA, Valle D, Schroer TA, Gould SJ. Expression of PEX11β mediates peroxisome proliferation in the absence of extracellular stimuli. J Biol Chem 1998; 273:29607–14; PMID:9792670; https://doi.org/10.1074/jbc.273.45.29607
- Schrader TA, Islinger M, Schrader M. Detection and immunolabeling of peroxisomal proteins. Methods Mol Biol 2017; 1595:113–130; PMID:28409457; https://doi.org/10.1007/978-1-4939-6937-1_12
- Palumbo V, Pellacani C, Heesom KJ, Rogala KB, Deane CM, Mottier-Pavie V, Gatti M, Bonaccorsi S, Wakefield JG. Misato controls mitotic microtubule generation by stabilizing the tubulin chaperone protein-1 complex. Curr Biol 2015; 25:1777–83; PMID:26096973; https://doi.org/10.1016/j.cub.2015.05.033
- Schuldiner M, Zalckvar E. Incredibly close—A newly identified peroxisome-ER contact site in humans. J Cell Biol 2017; 216:287–9; PMID:28108527
- Hua R, Cheng D, Coyaud E, Freeman S, Di Pietro E, Wang Y, Vissa A, Yip CM, Fairn GD, Braverman N, et al. VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J Cell Biol 2017; 216:367–77; PMID:28108526
- Abu-Safieh L, Alrashed M, Anazi S, Alkuraya H, Khan AO, Al-Owain M, Al-Zahrani J, Al-Abdi L, Hashem M, Al-Tarimi S, et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res 2013; 23:236–47; PMID:23105016; https://doi.org/10.1101/gr.144105.112
- Ferdinandusse S, Falkenberg K, Koster J, Mooyer P, Jones R, van Roermund C, Pizzino A, Schrader M, Wanders R, Vanderver A, et al. ACBD5 deficiency causes a defect in peroxisomal very long-chain fatty acid metabolism. J Med Genet 2016; pii: jmedgenet-2016-104132 [Epub ahead of print], PMID: 27799409; https://doi.org/10.1136/jmedgenet-2016-104132
- Yagita Y, Shinohara K, Abe Y, Nakagawa K, Al-Owain M, Alkuraya FS, Fujiki Y. Deficiency of a Retinal Dystrophy Protein, Acyl-CoA Binding Domain-containing 5 (ACBD5), Impairs Peroxisomal β-oxidation of very-long-chain fatty acids. J Biol Chem 2017; 292:691–705; PMID:27899449; https://doi.org/10.1074/jbc.M116.760090
- Baker A, Carrier DJ, Schaedler T, Waterham HR, Roermund CW, Theodoulou FL. Peroxisomal ABC transporters: functions and mechanism. Biochem Soc Trans 2015; 43:959–65; PMID:26517910; https://doi.org/10.1042/BST20150127
- van Roermund CWT, Visser WF, Ijlst L, van Cruchten A, Boek M, Kulik W, Waterham HR, Wanders RJA. The human peroxisomal ABC half transporter ALDP functions as a homodimer and accepts acyl-CoA esters. FASEB J 2008; 22:4201–8; PMID:18757502; https://doi.org/10.1096/fj.08-110866
- Geillon F, Gondcaille C, Raas Q, Dias AMM, Pecqueur D, Truntzer C, Lucchi G, Ducoroy P, Falson P, Savary S, et al. Peroxisomal ATP-binding cassette transporters form mainly tetramers. J Biol Chem 2017
- Yang IS, Son H, Kim S, Kim S. {ISOexpresso}: a web-based platform for isoform-level expression analysis in human cancer. BMC Genomics 2016; 17:631; PMID:27519173; https://doi.org/10.1186/s12864-016-2852-6
- Prinz WA. Bridging the gap : Membrane contact sites in signaling, metabolism, and organelle dynamics. 2014; 205:759–69
- Bonekamp NA, Vormund K, Jacob R, Schrader M. Dynamin-like protein 1 at the Golgi complex: a novel component of the sorting/targeting machinery en route to the plasma membrane. Exp Cell Res 2010; 316:3454–67; PMID:20688057; https://doi.org/10.1016/j.yexcr.2010.07.020
- Bonekamp NA, Islinger M, Lázaro MG, Schrader M. Cytochemical detection of peroxisomes and mitochondria. Methods Mol Biol 2013; 931:467–82; PMID:23027018
