ABSTRACT
Nucleotide-binding domain, leucine-rich-repeat–containing proteins (NLRs) are intracellular innate immune sensors of pathogen-associated and damage-associated molecular patterns. NLRs regulate diverse biologic processes such as inflammatory responses, cell proliferation and death, and gut microbiota to attenuate tumorigenesis. In a recent publication in Nature, we identified NLRC3 as a negative regulator of PI3K–mTOR signaling and characterized its potential tumor suppressor function. Enterocytes lacking NLRC3 cannot control cellular proliferation because they are unable to suppress activation of PI3K–mTOR signaling pathways. In this Extra-View, we explore possible mechanisms through which NLRC3 regulates cellular proliferation and cell death. Besides interacting with PI3K, NLRC3 associates with TRAF6 and mTOR, confirming our recent finding that NLRC3 negatively regulates the PI3K–mTOR axis. Herein, we show that NLRC3 suppresses c-Myc expression and activation of PI3K–AKT targets FoxO3a and FoxO1 in the colon of Nlrc3−/− mice, suggesting that additional signaling pathways contribute to increased cellular proliferation. Moreover, NLRC3 suppresses colorectal tumorigenesis by promoting cellular apoptosis. Genes encoding intestinal stem cell markers BMI1 and OLFM4 are upregulated in the colon of Nlrc3−/− mice. Herein, we discuss recent findings and explore mechanisms through which NLRC3 regulates PI3K–mTOR signaling. Our studies highlight the therapeutic potential of modulating NLRC3 to prevent and treat cancer.
Introduction
Nucleotide-binding domain (NOD), leucine-rich-repeat (LRR)–containing proteins (NLRs) are a large family of cytoplasmic sensors that regulate an extraordinarily diverse range of biologic functions. Deregulation of the functional activity of NLRs leads to the development of inflammatory diseases, autoimmunity and reproductive diseases.Citation1,2 Certain NLRs play a role in the initiation of inflammation, whereas others mediate the suppression of inflammation. Therefore, the coordinated activation and suppression of NLRs is essential to overcome infection and physiologic aberration and to restore homeostasis. NLRP3, NLRP6, NLRP12, NLRC4, and the DNA sensor AIM2 have central roles in preventing intestinal inflammation and colorectal cancer (CRC).Citation3-11
NLRC3 (also known as CLR16.2 or NOD3) is a poorly characterized member of the NLR family.Citation1 It was identified in a genomic screen of genes encoding proteins bearing LRRs and nucleotide-binding domains.Citation12 NLRC3 consists of an N-terminal caspase activation and recruitment domain, a central nucleotide-binding domain, and a C-terminal LRR domain.Citation13,14 NLRC3 is highly expressed in human and mouse immune cells.Citation14 Luciferase reporter assays show that overexpression of NLRC3 in 293T and Jurkat T cells impairs the activation of NF-κB.Citation14 NLRC3 functions as a negative regulator of NF-κB activation downstream of Toll-like receptors (TLRs)Citation13 and type I interferon production downstream of the stimulator of interferon genes (STING), a DNA sensor.Citation15 A meta-analysis recently identified a potential link between NLRC3 and cancer.Citation16 An analysis of 10 databases revealed lower expression of the gene encoding NLRC3 in tumors of patients with CRC than of healthy controls,Citation16 highlighting a potential role for NLRC3 in the development of CRC. Our recent study shows that NLRC3 is critical in restricting tumorigenesis in both a colitis-associated CRC model and the ApcMin/+ model, a spontaneous mouse model of intestinal cancer. Further, enterocytes lacking NLRC3 cannot control cellular proliferation because of a failure in suppressing the activation of phosphatidylinositol-3-kinase (PI3K) – mechanistic target of rapamycin (mTOR) signaling axis.Citation17 NLRC3 inhibits association of the PI3K p85 subunit with the PI3K p110α catalytic subunit, thereby preventing the activation of PI3K and its downstream targets PDK1, AKT, and mTOR.Citation17 Our new finding of the direct association of NLRC3 with TRAF6 and mTOR provides an additional molecular mechanism by which NLRC3 regulates mTOR modulation. Apart from mTOR activation, NLRC3 also regulated cellular proliferation by suppressing c-Myc expression and reducing the activation of PI3K–AKT targets, the forkhead box O (FoxO) proteins FoxO3a and FoxO1. Increased expression of genes encoding BMI1 and OLFM4 in the colon of mice lacking NLRC3 suggests that NLRC3 regulates proliferative signaling pathways involved in stem cell proliferation, maintenance, and differentiation. Moreover, the reduced activation of caspase-8, caspase-7 and caspase-3 in colon of Nlrc3−/− mice suggests that the tumor suppressor function of NLRC3 is also attributed to its ability to promote apoptosis. From the clinical standpoint, understanding the complex network through which NLRC3 promotes intestinal homeostasis can lead to the development of new therapeutic approaches to prevent and treat cancer.
Results and discussion
NLRC3 sequesters TRAF6 to modulate mTOR signaling
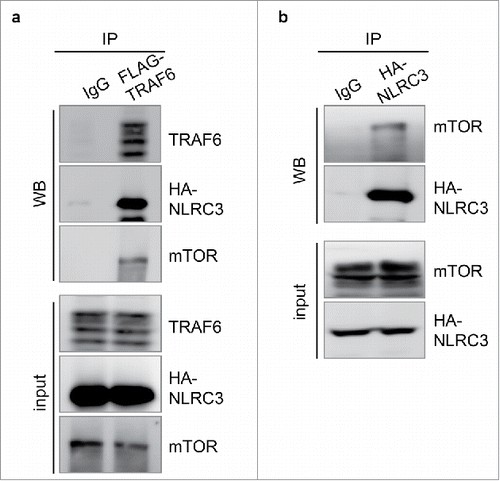
In our recent study, we reported that deletion of NLRC3 results in the co-localization of mTOR with lysosomal-associated membrane protein 1, leading to auto-activation of mTOR, which subsequently targets S6 and 4E-BP1.Citation17 The interaction of NLRC3 with PI3K subunits, but not with AKT and PDK1, inhibits activation of the PI3K–AKT–mTOR axis.Citation17 NLRC3 contains binding sites for tumor necrosis factor receptor–associated factors (TRAFs) within the nucleotide-binding domain, which allow its association with the TLR signaling molecule TRAF6.Citation13 Association with NLRC3 leads to auto-ubiquitylation and degradation of TRAF6, thereby tempering TLR-dependent activation of the NF-κB signaling pathway.Citation13 Similarly, the interaction of TRAF6 with p62 is required for translocation of mTOR to the lysosome for its subsequent activation.Citation18 One possibility is that NLRC3 regulates mTOR activation by directly associating with TRAF6 and mTOR. To investigate this, we overexpressed NLRC3, TRAF6, and mTOR in 293T cells and co-immunoprecipitated with TRAF6. We observed that NLRC3, TRAF6, and mTOR were in the same complex (). Consistent with this, NLRC3-based immunoprecipitation also demonstrated the interaction of NLRC3 with mTOR, further establishing that NLRC3 plays a role in the mTOR signaling pathway (). Given that NLRC3 interacts with TRAF6 and leads to its degradation,Citation13 loss of NLRC3 provides the TRAF6–p62 complex a platform to translocate mTOR to the lysosome for its subsequent activation. In line with our recent observation that loss of NLRC3 results in higher cellular proliferation owing to increased activation of mTOR signaling,Citation17 another study reported that loss of TRAF6 impairs division in cancer cellsCitation18 due to defective activation of mTOR.
Figure 1. NLRC3 sequesters mTOR-TRAF6 complex. (A) Human embryonic kidney (HEK) 293T cells were transfected with the FLAG-TRAF6, HA-NLRC3, and mTOR plasmids. TRAF6 immunoprecipitates were analyzed for TRAF6, HA, and mTOR expression by western blot. (B) 293T cells were transfected with HA-NLRC3 and mTOR plasmids. HA immunoprecipitates were analyzed for mTOR and HA expression. Data represent 1 experiment representative of 2 independent experiments.
NLRC3 does not regulate inflammasome activation
Certain members of the NLR family can form inflammasome complexes.Citation19 Inflammasome-mediated caspase-1 activation during dextran sulfate sodium (DSS)–induced colitis is protective against colitis and colitis-associated tumorigenesis, particularly through the production of IL-18.Citation3,4,9,20 However, we found that caspase-1 was similarly activated in wild-type (WT) and Nlrc3−/− mice (), which is in line with our previous finding that IL-18 production is similar in colons of WT and Nlrc3−/− mice 14 d after azoxymethane (AOM) injection.Citation17
Figure 2. NLRC3 regulates cellular proliferation and apoptosis. (A) Immunoblot analysis of caspase-1 (CASP-1), caspase-8 (CASP-8), caspase-3 (CASP-3), and caspase-7 (CASP-7) activation and GAPDH (loading control) in the colon of WT and Nlrc3−/− mice 14 d after AOM injection. (B) Immunoblot analysis of c-Myc, phosphorylated FoxO3a and FoxO1 (P-FoxO3a and P-FoxO1), and GAPDH (loading control) in the colon of WT and Nlrc3−/− mice 14 d after AOM injection. Protein band intensity was normalized to the loading control and expressed relative to that of the WT, set at 1. (C) Images and quantification of the number of cyclin D1+ cells in each crypt of WT (day 0, n = 5; day 14, n = 8) and Nlrc3−/− (day 0, n = 5; day 14, n = 8) mice (left). Scale bar, 200 μm (C). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not statistically significant [2-tailed t-test (B and C)]. Data are from 1 experiment representative of 2 (mean and s.e.m. in A and C).
![Figure 2. NLRC3 regulates cellular proliferation and apoptosis. (A) Immunoblot analysis of caspase-1 (CASP-1), caspase-8 (CASP-8), caspase-3 (CASP-3), and caspase-7 (CASP-7) activation and GAPDH (loading control) in the colon of WT and Nlrc3−/− mice 14 d after AOM injection. (B) Immunoblot analysis of c-Myc, phosphorylated FoxO3a and FoxO1 (P-FoxO3a and P-FoxO1), and GAPDH (loading control) in the colon of WT and Nlrc3−/− mice 14 d after AOM injection. Protein band intensity was normalized to the loading control and expressed relative to that of the WT, set at 1. (C) Images and quantification of the number of cyclin D1+ cells in each crypt of WT (day 0, n = 5; day 14, n = 8) and Nlrc3−/− (day 0, n = 5; day 14, n = 8) mice (left). Scale bar, 200 μm (C). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not statistically significant [2-tailed t-test (B and C)]. Data are from 1 experiment representative of 2 (mean and s.e.m. in A and C).](/cms/asset/c1ea7116-1571-4d58-a4f4-ba14588e9178/kccy_a_1317414_f0002_c.gif)
NLRC3 regulates cellular proliferation and apoptosis
Alterations in the physiologic equilibrium between epithelial proliferation and apoptosis in the colonic mucosa are associated with increased risk of CRC. The loss of equilibrium occurs in the very early phases of tumorigenesis.Citation21 Since alteration in the signaling molecules associated with epithelial proliferation and apoptosis in response to AOM/DSS peaks in colitic phase, which happens 14 d after AOM injection,Citation17 we used this time point for further analyses. Mice lacking NLRC3 have increased numbers of Ki67+ and PCNA+ cells in the intestinal crypt, suggesting increased proliferation of the colonic epithelium.Citation17 Multiple signaling pathways can regulate cellular proliferation. Apart from the regulation of mTOR targets, negative regulation by NLRC3 at the PI3K–AKT–mTOR axis can affect the expression or regulation of various molecules critical in cellular proliferation. Emerging evidences suggest that PI3K–AKT–mTOR signaling regulates transcriptional activity of c-Myc,Citation22,23 which is frequently deregulated in various cancers, including gastrointestinal cancers.Citation24 We found that mice lacking NLRC3 had increased expression of the oncoprotein c-Myc in the colon 14 d after AOM injection (). c-Myc is required for the transcription of many cell cycle–related proto-oncogenes such as cyclin D1,Citation25 and cyclin D1 is overexpressed in various human cancers, including colon cancer.Citation26 We identified cyclin D1+ cells in the intestinal epithelium by immunohistochemical analysis. Before exposure to AOM and DSS, numbers of cyclin D1+ cells were similar in the intestinal epithelium of WT mice and Nlrc3−/− mice (). However, 14 d after AOM injection, numbers of cyclin D1+ cells per intestinal crypt were significantly higher in Nlrc3−/− mice than in WT mice ().
Besides activating mTOR, the PI3K–AKT signaling axis phosphorylates FoxO proteins FoxO3a and FoxO1, which are considered bona fide tumor suppressors because they regulate cell proliferation, apoptosis, metabolism, and survival.Citation27 Deregulation of genes encoding FoxO3a and FoxO1 is associated with the progression of several types of tumors, including CRC.Citation28-30 AKT-mediated phosphorylation leads to sequestration of FoxO3a and FoxO1 in the cytosol in an inactive state and ubiquitin-mediated degradation.Citation31 Because NLRC3 regulates the PI3K–AKT signaling axis, we determined the phosphorylation status of FoxO3a and FoxO1 in colons of WT and Nlrc3−/− mice 14 d after AOM injection. Consistent with increased activation of AKT, we observed higher phosphorylation of FoxO3a and FoxO1 in the colon of Nlrc3−/− mice than of WT mice (). These results support that FoxO3a and FoxO1 are potential targets of NLRC3 for regulation of tumorigenesis.
Reduced apoptosis can also contribute to increased tumor burden. Therefore, we determined caspase-8, caspase-3, and caspase-7 activation in colons of WT and Nlrc3−/− mice 14 d after AOM injection. Caspase-8, caspase-3, and caspase-7 activation was lower in the colon of Nlrc3−/− mice than of WT mice (), suggesting that reduced apoptosis during the early phase in the absence of NLRC3 contributes to increased tumorigenesis. However, the mechanism by which NLRC3 promotes cell death remains to be determined.
NLRC3 regulates colonic stem cells and sensitivity to tumorigenesis
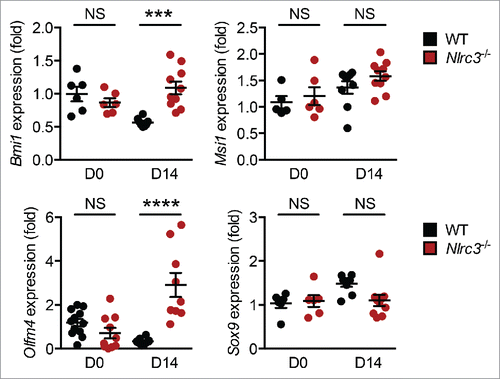
Stem cells are considered to be the cells of origin of CRC because they can undergo self-renewal, unlimited proliferation, and differentiation.Citation32 Several markers, such as LRR-containing G-protein–coupled receptor 5 (LGR5), sex-determining region y-box 9 (SOX9), olfactomedin-4 (OLFM4), and Musashi1 (MSI1), have been used to identify stem cells and quantify their stemness.Citation32,33 Colonic stem cells harvested from mice lacking NLRC3 develop more readily into organoids than do those isolated from WT mice.Citation17 Expression of genes encoding stem cell markers LGR5,Citation17 BMI1, MSI1, OLFM4, and SOX9 was similar in colon tissues from WT and Nlrc3−/− mice () before AOM injection, suggesting that the increased number and size of intestinal organoids derived from the colonic epithelium of Nlrc3−/− mice were because of increased colony-forming capacity rather than differences in the numbers of starting intestinal stem cells. There was no change in the expression of Lgr5, Msi1, and Sox9 in the colons of WT and Nlrc3−/− mice 14 d after AOM injection (ref. Citation17 and ). However, the expression of genes encoding BMI1 and OLFM4 was significantly higher in the colon of Nlrc3−/− mice than in WT mice 14 d after AOM injection (). Aberrant expression of BMI1 has been associated with increased risk of colon cancer, and BMI1 inhibition by small molecules reduces tumor burden in primary human colorectal tumor xenograft models.Citation34 Unlike Lgr5+ cells, the Bmi1+ population is relatively quiescent, radiation resistant, and can regenerate in response to injury or ablation of Lgr5-expressing cells.Citation35 Given that the PI3K–AKT signaling axis mediates the phosphorylation of BMI1 and enhances its ability to promote prostate carcinogenesis,Citation36 it is possible that NLRC3 suppresses PI3K-AKT–mediated BMI1 phosphorylation to inhibit colorectal tumorigenesis.
Figure 3. NLRC3 regulates stem cell markers. Relative expression of genes encoding BMI1, MSI1, OLFM4, and SOX9 in colon tissues of untreated WT and Nlrc3−/− mice or in WT and Nlrc3−/− mice 14 d after AOM injection. Each symbol represents 1 mouse. ***P < 0.001; ****P < 0.0001; NS, not statistically significant (2-tailed t-test). Data represent 2 independent experiments (mean and s.e.m.).
Similarly, expression of the gene encoding OLFM4 is upregulated in human inflammatory bowel diseaseCitation37 and early-stage colon cancer.Citation38 Transcription of the gene encoding OLFM4 is regulated by NF-kB,Citation39 Notch,Citation40 and PU.1.Citation41 The negative regulatory role of NLRC3 in NF-κB signaling downstream of TLRs can at least explain the increased expression of Olfm4 in the colon of Nlrc3−/− mice. Whether NLRC3 also regulates Notch and PU.1 remains to be established. Moreover, inhibition of PI3K activation is sufficient to suppress OLFM4 expression,Citation39 which could explain decreased Olfm4 expression in the colon of Nlrc3−/− mice. OLFM4 interacts with NLRC1 and NLRC2,Citation42 which are intracellular bacterial sensors associated with Crohn disease.Citation43,44 Future studies are needed to investigate the interaction between NLRC3 and OLFM4.
Since activation of the PI3K–AKT pathway positively correlates with upregulation of Bmi1 and Olfm4 expression in the colon of Nlrc3−/− mice, studying the post-translational modifications in BMI1 and OLFM4 is likely to provide mechanistic insights into how their oncogenic potential can be manipulated.
Colorectal cancer in Nlrc3−/− mice is not driven by microbiota
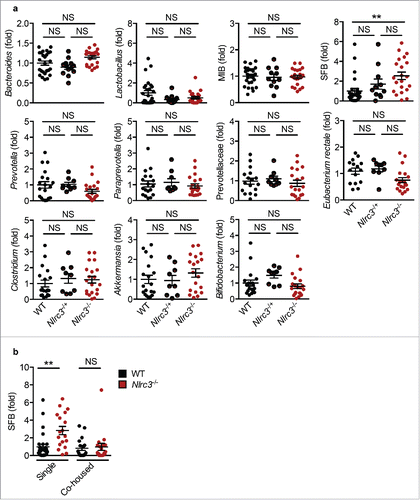
The human gut is colonized by a diverse microbial population that includes bacteria, fungi and viruses,Citation45 which play a pivotal role in maintaining homeostasis in the gut. Breakdown of the homeostasis by dysbiosis or deregulation of immune responses results in various pathological conditions such as inflammatory bowel diseases and CRC.Citation46 To reduce the occurrence of such pathological conditions, the gut has several innate immune sensors belonging to the NLR family that are critical for modulating microbial ecology to protect against tumor development.Citation47 Intestinal cell proliferation and progression of CRC is regulated by the composition of gut microbiota. To investigate whether an altered gut microbiota increases the susceptibility of Nlrc3−/− mice to CRC, we analyzed the levels of major bacterial species of gut microbiota in stool samples of separately housed (single housed) WT, Nlrc3+/–, and Nlrc3−/− mice. Real-time quantitative PCR of 11 major bacterial populations revealed that WT, Nlrc3+/–, and Nlrc3−/− mice harbored similar levels of Bacteroides, Lactobacillus, mouse intestinal Bacteroides (MIB), Prevotellaceae, Prevotella, Paraprevotella, Bifidobacterium, Eubacterium rectale, Clostridium, and Akkermansia. Compared with WT mice, Nlrc3−/− mice had elevated levels of segmented filamentous bacteria (SFB) (), which are associated with IL-17– and IL-22–mediated chronic inflammation.Citation48 Interestingly, co-housing equilibrated the relative abundance of SFB in WT and Nlrc3−/− mice (). However, Nlrc3−/− mice co-housed with WT mice were as susceptible to tumorigenesis as were separately housed Nlrc3−/− mice.Citation17 These findings suggest that differences in these bacterial populations do not contribute to the protective role of NLRC3 during the development of CRC.
Figure 4. Landscape of gut microbiota in Nlrc3−/− mice. (A) Levels of Bacteroides, Lactobacillus, mouse intestinal Bacteroides (MIB), segmented filamentous bacteria (SFB), Prevotella, Paraprevotella, Prevotellaceae, Eubacterium rectale, Clostridium, Akkermansia, and Bifidobacterium in untreated WT, Nlrc3−/+ and Nlrc3−/− mice. (B) Levels of SFB in separately housed (single housed) WT and Nlrc3−/− mice and co-housed WT and Nlrc3−/− mice. The level of genes was expressed relative to that of the WT, set at 1. Each symbol represents 1 mouse. **P < 0.01; NS, not statistically significant (2-tailed t-test). Data represent 2 independent experiments (mean and s.e.m.).
Future directions
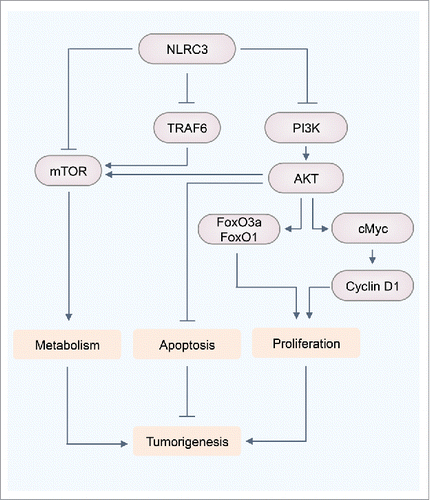
Our current and previously published studies have established that NLRC3 negatively regulates PI3K–mTOR pathways, which integrate upstream signals from growth factors, nutrients, and cellular energy to regulate various biologic processes involved in cell metabolism, growth, proliferation, and survival ().Citation17,49 Still, the role of NLRC3 in various other signaling pathways that drive cellular proliferation remains to be explored. The Wnt signaling pathway, which has been well studied in enterocyte proliferation, activates mTOR,Citation50 suggesting a link between NLRC3 and the Wnt signaling pathway. However, the exact mechanism by which NLRC3 modulates Wnt signaling needs further investigation. Changes in metabolite levels can contribute to tumorigenesis.Citation51 Thus, it is possible that NLRC3 acts as a nutrient sensor for the mTOR signaling pathway. Metabolic sensors such as AMP-activated protein kinase act as tumor suppressors by regulating mTOR activity and thus inhibiting the translation of many proteins required for rapid cell growth.Citation52 Hence, it is possible that there is crosstalk between these 2 metabolic sensors to regulate mTOR activity and suppress tumorigenesis. NLRC3-interacting partners mTOR and TRAF6 inhibit autophagy, a catabolic machinery to generate nutrients and energy required for cellular activities upon nutrient starvation,Citation53,54 but the exact mechanism of this regulation remains to be elucidated. The role of NLRC3 is not confined to cellular proliferation. Given the contribution of mTOR signaling to metabolic diseases such as diabetes, obesity, and atherosclerosis, and other inflammatory disorders such as arthritis,Citation55,56 a protective role of NLRC3 can be speculated in the treatment and prevention of these diseases. Our study and those by others demonstrated that NLRC3 contributes to the negative regulation of inflammation.Citation13,15,17 Moreover, the expression of NLRC3 is substantially downregulated in the nasal mucosa of patients with the autoimmune disease Wegener granulomatosis compared with that in healthy individuals.Citation57 Unraveling the molecular and metabolic responses modulated by NLRC3 could provide precious insights into developing therapeutics for infectious disease, autoinflammation, and cancer.
Methods
Cell culture
The embryonic kidney epithelial cell line HEK293T (ATCC#3216, American Type Culture Collection) was cultured in DMEM (11995073, ThermoFisher Scientific) supplemented with 10% fetal bovine serum (TMS-013-B, Millipore) and 1% penicillin and streptomycin (15070–063, ThermoFisher Scientific). Cells were maintained at 37°C with 5% CO2.
Mice
WT (C57BL/6) and Nlrc3–/– 17 mice were bred and maintained under specific pathogen-free conditions at St. Jude Children's Research Hospital (St. Jude), Memphis, TN. Animal study protocols were approved by the St. Jude Animal Care and Use Committee.
AOM-DSS model of colorectal tumorigenesis
Previously established protocols were followed to induce colitis-associated CRC in mice.Citation17
Western blotting
Proteins from the colon were extracted using RIPA lysis buffer supplemented with proteinase and phosphatase inhibitors (Roche). Western blotting was performed as described previously.Citation58 Primary antibodies were caspase-1 p10 (1:500 dilution, sc-515, Santa Cruz Biotechnology), c-Myc (1:1,000, #5605, Cell Signaling Technology), caspase-3 (1:1,000, #9662, Cell Signaling Technology), cleaved caspase-3 (1:1,000, #9661, Cell Signaling), caspase-7 (1:1,000, #9492, Cell Signaling Technology), cleaved caspase-7 (1:1,000, #9491, Cell Signaling), caspase-8 (1:1,000, #9746, Cell Signaling Technology), cleaved caspase-8 (1:1,000, #8592, Cell Signaling Technology), Phospho-FoxO1 and FoxO3a (1:1,000, #9464, Cell Signaling Technology), mTOR (1:1,000, 2972, Cell Signaling Technology), TRAF6 (1:1,000, #8028, Cell Signaling Technology), and GAPDH (1:10,000; #5174, Cell Signaling Technology). Immunoblots were quantified using Image J.
Histology and microscopy
Colons were processed as described previously.Citation58 Cyclin D1 expression in the intestinal epithelium was detected by immunoperoxidase staining for cyclin D1 (#241R-15, Cell Marque). The number of cyclin D1+ cells per crypt in each animal was counted (at least 18–20 crypts per mouse).
Quantitative real-time PCR
RNA was isolated from the colon by using Trizol (15596026, ThermoFisher Scientific) and reverse transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (4368814, Applied Biosystems). For PCR analysis of intestinal bacteria, fecal DNA was extracted as described previously.Citation59 Gene expression was assessed using the 2 × SYBR Green Master Mix (4368706, Applied Biosystems) according to the manufacturer's instructions. Sequences for qRT-PCR primers were as follows: Bmi1-F 5′-GTT CGA TGC ATT TCT GCT TG-3′; Bmi1-R 5-′TGG CTC GCA TTC ATT TTA TG-3′; Sox9-F 5′-TCC ACG AAG GGT CTC TTC TC-3′; Sox9-R 5′-AGG AAG CTG GCA GAC CAG TA-3′; Msi1-F 5′-AAT TCG GGG AAC TGG TAG GT-3′; Msi1-R 5′-GAT GCC TTC ATG CTG GGT AT-3′; Olmf4-F 5′-CAG CCA CTT TCC AAT TTC ACT G-3′; Olmf4-R 5′-GCT GGA CAT ACT CCT TCA CCT TA-3′; β-actin-F 5′-CAG CTT CTT TGC AGC TCC TT-3′, and β-actin-R 5′-CAC GAT GGA GGG GAA TAC AG-3′; Eubacteria (Universal)-F 5′-ACT CCT ACG GGA GGC AGC AGT-3′; Eubacteria (Universal)-R 5′-ATT ACC GCG GCT GCT GGC-3′; Prevotellaceae-F 5′-CCA GCC AAG TAG CGT GCA-3′; Prevotellaceae-R 5′-TGG ACC TTC CGT ATT ACC-3′; Bacteroides-F 5′-GGT TCT GAG AGG AGG TCC C-3′; Bacteroides-R 5′-GCT GCC TCC CGT AGG AGT-3′; MIB-F 5′-CCA GCA GCC GCG GTA ATA-3′; MIB-R 5′-CGC ATT CCG CT ACT TCT C-3′; SFB-F 5′-GAC GCT GAG GCA TGA GAG CT-3′; SFB-R 5′-GAC GGC ACG GAT TGT TAT TCA-3′; Lactobacillus-F 5′-GGA AAC AGA TGC TAA TAC CG-3′; Lactobacillus-R 5′-CAC CGC TAC ACA TGG AG-3′; Prevotella-F 5′-CAC GGT AAA CGA TGG ATG CC-3′; Prevotella-R 5′-GGT CGG GTT GCA GAC C-3′; Paraprevotella-F 5′-AGG GGC AGC ATG GAC CC-3′; Paraprevotella-R 5′-CCT TTC AGG AGA CTA TCC CGG A-3′; Clostridium-F 5′-CTC AAC TTG GGT GCT GCA TTT-3′; Clostridium-R ATT GTA GTA CGT GTG TAG CCC-5′; Akkermansia-F 5′-CAG CAC GTG AAG GTG GGG AC-3′; Akkermansia-R 5′-CCT TGC GGT TGG CTT CAG AT-3′; Eubacterium rectale-F 5′-GCT TCT TAG TCA GGT ACC GTC A-3′; Eubacterium rectale-R 5′-ACT CCT ACG GGA GGC AGC-3′; Bifidobacterium-F 5′-TCG CGT CYG GTG TGA AAG-3′; Bifidobacterium-R 5′-CCA CT CCA GCR TCC AC-3′. Levels of Bmi1, Msi1, Olfm4, and Sox9 expression were normalized to β-actin. Levels of the 16S rRNA gene from each bacterial population were normalized to the 16S rRNA gene of Eubacteria.
Co-immunoprecipitation assays
Co-immunoprecipitation assays were performed as described previously.Citation17
Statistical analyses
GraphPad Prism 6.0 software was used for data analysis. Data are shown as mean ± standard error of the mean (s.e.m.). Statistical significance was determined by t tests (2-tailed) for 2 groups or one-way ANOVA (with multiple comparisons tests) for 3 or more groups. P < 0.05 was considered statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
R.K. and T.D.K. conceptualized the study; R.K. and R.K.S.M. designed the methodology; R.K., R.K.S.M., and Q.Z. performed the experiments; R.K., R.K.S.M., and Q.Z. conducted the analysis; R.K. and T.D.K. wrote the original draft of the manuscript; R.K.S.M. and Q.Z. reviewed and edited the manuscript; and T.D.K. provided the resources and supervised the study.
Acknowledgments
We thank scientific editing department at St. Jude for their comments and suggestions.
Funding
Work from our laboratory is supported by the US National Institutes of Health (AI101935, AI124346, AR056296, and CA163507 to T.D.K.) and ALSAC (to T.D.K.).
References
- Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity 2007; 27:549-59; PMID:17967410; https://doi.org/10.1016/j.immuni.2007.10.002
- Van Gorp H, Kuchmiy A, Van Hauwermeiren F, Lamkanfi M. NOD-like receptors interfacing the immune and reproductive systems. FEBS J 2014; 281:4568-82; PMID:25154302; https://doi.org/10.1111/febs.13014
- Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 2010; 32:379-91; PMID:20303296; https://doi.org/10.1016/j.immuni.2010.03.003
- Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol 2010; 185:4912-20; PMID:20855874; https://doi.org/10.4049/jimmunol.1002046
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011; 145:745-57; PMID:21565393; https://doi.org/10.1016/j.cell.2011.04.022
- Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RK, Gurung P, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell 2015; 162:45-58; PMID:26095253; https://doi.org/10.1016/j.cell.2015.06.001
- Hu B, Elinav E, Huber S, Booth CJ, Strowig T, Jin C, Eisenbarth SC, Flavell RA. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci U S A 2010; 107:21635-40; PMID:21118981; https://doi.org/10.1073/pnas.1016814108
- Allen IC, Wilson JE, Schneider M, Lich JD, Roberts RA, Arthur JC, Woodford RM, Davis BK, Uronis JM, Herfarth HH, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity 2012; 36:742-54; PMID:22503542; https://doi.org/10.1016/j.immuni.2012.03.012
- Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med 2010; 207:1045-56; PMID:20385749; https://doi.org/10.1084/jem.20100050
- Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 2014; 156:1045-59; PMID:24581500; https://doi.org/10.1016/j.cell.2014.01.026
- Wilson JE, Petrucelli AS, Chen L, Koblansky AA, Truax AD, Oyama Y, Rogers AB, Brickey WJ, Wang Y, Schneider M, et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med 2015; 21:906-13; PMID:26107252; https://doi.org/10.1038/nm.3908
- Harton JA, Linhoff MW, Zhang J, Ting JP. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol 2002; 169:4088-93; PMID:12370334; https://doi.org/10.4049/jimmunol.169.8.4088
- Schneider M, Zimmermann AG, Roberts RA, Zhang L, Swanson KV, Wen H, Davis BK, Allen IC, Holl EK, Ye Z, et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol 2012; 13:823-31; PMID:22863753; https://doi.org/10.1038/ni.2378
- Conti BJ, Davis BK, Zhang J, O'Connor W, Jr., Williams KL, Ting JP. CATERPILLER 16.2 (CLR16.2), a novel NBD/LRR family member that negatively regulates T cell function. J Biol Chem 2005; 280:18375-85; PMID:15705585; https://doi.org/10.1074/jbc.M413169200
- Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM, Zhang Z, Schneider M, Jiang Y, Fitzgerald KA, et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 2014; 40:329-41; PMID:24560620; https://doi.org/10.1016/j.immuni.2014.01.010
- Liu R, Truax AD, Chen L, Hu P, Li Z, Chen J, Song C, Chen L, Ting JP. Expression profile of innate immune receptors, NLRs and AIM2, in human colorectal cancer: correlation with cancer stages and inflammasome components. Oncotarget 2015; 6:33456-69; PMID:26378020
- Karki R, Man SM, Malireddi RK, Kesavardhana S, Zhu Q, Burton AR, Sharma BR, Qi X, Pelletier S, Vogel P, et al. NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature 2016; 540:583-587; PMID:27951586; https://doi.org/10.1038/nature20597
- Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell 2013; 51:283-96; PMID:23911927; https://doi.org/10.1016/j.molcel.2013.06.020
- Man SM, Kanneganti TD. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat Rev Immunol 2016; 16:7-21; PMID:26655628; https://doi.org/10.1038/nri.2015.7
- Karki R, Man SM, Kanneganti TD. Inflammasomes and Cancer. Cancer Immunol Res 2017; 5(2):94-99; PMID:28093447
- Anti M, Armuzzi A, Morini S, Iascone E, Pignataro G, Coco C, Lorenzetti R, Paolucci M, Covino M, Gasbarrini A, et al. Severe imbalance of cell proliferation and apoptosis in the left colon and in the rectosigmoid tract in subjects with a history of large adenomas. Gut 2001; 48:238-46; PMID:11156647; https://doi.org/10.1136/gut.48.2.238
- Zhu J, Blenis J, Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci U S A 2008; 105:6584-9; PMID:18451027; https://doi.org/10.1073/pnas.0802785105
- Sander S, Calado DP, Srinivasan L, Kochert K, Zhang B, Rosolowski M, Rodig SJ, Holzmann K, Stilgenbauer S, Siebert R, et al. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 2012; 22:167-79; PMID:22897848; https://doi.org/10.1016/j.ccr.2012.06.012
- Wang C, Lisanti MP, Liao DJ. Reviewing once more the c-myc and Ras collaboration: converging at the cyclin D1-CDK4 complex and challenging basic concepts of cancer biology. Cell Cycle 2011; 10:57-67; PMID:21200143; https://doi.org/10.4161/cc.10.1.14449
- Obaya AJ, Mateyak MK, Sedivy JM. Mysterious liaisons: the relationship between c-Myc and the cell cycle. Oncogene 1999; 18:2934-41; PMID:10378690; https://doi.org/10.1038/sj.onc.1202749
- Wang QS, Papanikolaou A, Sabourin CL, Rosenberg DW. Altered expression of cyclin D1 and cyclin-dependent kinase 4 in azoxymethane-induced mouse colon tumorigenesis. Carcinogenesis 1998; 19:2001-6; PMID:9855016; https://doi.org/10.1093/carcin/19.11.2001
- Chiacchiera F, Simone C. The AMPK-FoxO3A axis as a target for cancer treatment. Cell Cycle 2010; 9:1091-6; PMID:20190568; https://doi.org/10.4161/cc.9.6.11035
- Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene 2008; 27:2312-9; PMID:18391973; https://doi.org/10.1038/onc.2008.24
- Tenbaum SP, Ordonez-Moran P, Puig I, Chicote I, Arques O, Landolfi S, Fernandez Y, Herance JR, Gispert JD, Mendizabal L, et al. beta-catenin confers resistance to PI3K and AKT inhibitors and subverts FOXO3a to promote metastasis in colon cancer. Nat Med 2012; 18:892-901; PMID:22610277; https://doi.org/10.1038/nm.2772
- Fu Q, Du Y, Yang C, Zhang D, Zhang N, Liu X, Cho WC, Yang Y. An oncogenic role of miR-592 in tumorigenesis of human colorectal cancer by targeting Forkhead Box O3A (FoxO3A). Expert Opin Ther Targets 2016; 20:771-82; PMID:27167185; https://doi.org/10.1080/14728222.2016.1181753
- Plas DR, Thompson CB. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem 2003; 278:12361-6; PMID:12517744; https://doi.org/10.1074/jbc.M213069200
- Fanali C, Lucchetti D, Farina M, Corbi M, Cufino V, Cittadini A, Sgambato A. Cancer stem cells in colorectal cancer from pathogenesis to therapy: controversies and perspectives. World J Gastroenterol 2014; 20:923-42; PMID:24574766; https://doi.org/10.3748/wjg.v20.i4.923
- Espersen ML, Olsen J, Linnemann D, Hogdall E, Troelsen JT. Clinical implications of intestinal stem cell markers in colorectal cancer. Clin Colorectal Cancer 2015; 14:63-71; PMID:25657049; https://doi.org/10.1016/j.clcc.2014.12.004
- Kreso A, van Galen P, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, Cao L, Baiazitov R, Du W, Sydorenko N, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med 2014; 20:29-36; PMID:24292392; https://doi.org/10.1038/nm.3418
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A 2012; 109:466-71; PMID:22190486; https://doi.org/10.1073/pnas.1118857109
- Nacerddine K, Beaudry JB, Ginjala V, Westerman B, Mattiroli F, Song JY, van der Poel H, Ponz OB, Pritchard C, Cornelissen-Steijger P, et al. Akt-mediated phosphorylation of Bmi1 modulates its oncogenic potential, E3 ligase activity, and DNA damage repair activity in mouse prostate cancer. J Clin Invest 2012; 122:1920-32; PMID:22505453; https://doi.org/10.1172/JCI57477
- Gersemann M, Becker S, Nuding S, Antoni L, Ott G, Fritz P, Oue N, Yasui W, Wehkamp J, Stange EF. Olfactomedin-4 is a glycoprotein secreted into mucus in active IBD. J Crohns Colitis 2012; 6:425-34; PMID:22398066; https://doi.org/10.1016/j.crohns.2011.09.013
- Besson D, Pavageau AH, Valo I, Bourreau A, Belanger A, Eymerit-Morin C, Mouliere A, Chassevent A, Boisdron-Celle M, Morel A, et al. A quantitative proteomic approach of the different stages of colorectal cancer establishes OLFM4 as a new nonmetastatic tumor marker. Mol Cell Proteomics 2011; 10:1-14; PMID:21986994; https://doi.org/10.1074/mcp.M111.009712
- Chin KL, Aerbajinai W, Zhu J, Drew L, Chen L, Liu W, Rodgers GP. The regulation of OLFM4 expression in myeloid precursor cells relies on NF-kappaB transcription factor. Br J Haematol 2008; 143:421-32; PMID:18764868; https://doi.org/10.1111/j.1365-2141.2008.07368.x
- VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud A, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 2012; 139:488-97; PMID:22190634; https://doi.org/10.1242/dev.070763
- Rosenbauer F, Wagner K, Zhang P, Knobeloch KP, Iwama A, Tenen DG. pDP4, a novel glycoprotein secreted by mature granulocytes, is regulated by transcription factor PU.1. Blood 2004; 103:4294-301; PMID:14962908; https://doi.org/10.1182/blood-2003-08-2688
- Liu W, Yan M, Liu Y, Wang R, Li C, Deng C, Singh A, Coleman WG, Jr., Rodgers GP. Olfactomedin 4 down-regulates innate immunity against Helicobacter pylori infection. Proc Natl Acad Sci U S A 2010; 107:11056-61; PMID:20534456; https://doi.org/10.1073/pnas.1001269107
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 2001; 411:603-6; PMID:11385577; https://doi.org/10.1038/35079114
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 2001; 411:599-603; PMID:11385576; https://doi.org/10.1038/35079107
- Gagniere J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, Bringer MA, Pezet D, Bonnet M. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol 2016; 22:501-18; PMID:26811603; https://doi.org/10.3748/wjg.v22.i2.501
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007; 448:427-34; PMID:17653185; https://doi.org/10.1038/nature06005
- Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 2013; 13:321-35; PMID:23618829; https://doi.org/10.1038/nri3430
- Russo E, Taddei A, Ringressi MN, Ricci F, Amedei A. The interplay between the microbiome and the adaptive immune response in cancer development. Therap Adv Gastroenterol 2016; 9:594-605; PMID:27366226; https://doi.org/10.1177/1756283X16635082
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci 2009; 122:3589-94; PMID:19812304; https://doi.org/10.1242/jcs.051011
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 2006; 126:955-68; PMID:16959574; https://doi.org/10.1016/j.cell.2006.06.055
- Sullivan LB, Gui DY, Vander Heiden MG. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat Rev Cancer 2016; 16:680-93; PMID:27658530; https://doi.org/10.1038/nrc.2016.85
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol 2003; 2:28; PMID:14511394; https://doi.org/10.1186/1475-4924-2-28
- Nazio F, Cecconi F. mTOR, AMBRA1, and autophagy: an intricate relationship. Cell Cycle 2013; 12:2524-5; PMID:23907135; https://doi.org/10.4161/cc.25835
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15:406-16; PMID:23524951; https://doi.org/10.1038/ncb2708
- Jia G, Aroor AR, Martinez-Lemus LA, Sowers JR. Overnutrition, mTOR signaling, and cardiovascular diseases. Am J Physiol Regul Integr Comp Physiol 2014; 307:R1198-206; PMID:25253086; https://doi.org/10.1152/ajpregu.00262.2014
- Cejka D, Hayer S, Niederreiter B, Sieghart W, Fuereder T, Zwerina J, Schett G. Mammalian target of rapamycin signaling is crucial for joint destruction in experimental arthritis and is activated in osteoclasts from patients with rheumatoid arthritis. Arthritis Rheum 2010; 62:2294-302; PMID:20506288; https://doi.org/10.1002/art.27504
- Laudien M, Hasler R, Wohlers J, Bock J, Lipinski S, Bremer L, Podschun R, Ambrosch P, Lamprecht P, Rosenstiel P, et al. Molecular signatures of a disturbed nasal barrier function in the primary tissue of Wegener's granulomatosis. Mucosal Immunol 2011; 4:564-73; PMID:21412229; https://doi.org/10.1038/mi.2011.9
- Karki R, Man SM, Malireddi RK, Gurung P, Vogel P, Lamkanfi M, Kanneganti TD. Concerted Activation of the AIM2 and NLRP3 Inflammasomes Orchestrates Host Protection against Aspergillus Infection. Cell host & microbe 2015; 17:357-68; https://doi.org/10.1016/j.chom.2015.01.006
- Zhu Q, Man SM, Gurung P, Liu Z, Vogel P, Lamkanfi M, Kanneganti TD. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal infla.mmation. J Immunol 2014; 193:4779-82; PMID:25320273; https://doi.org/10.4049/jimmunol.1402051