ABSTRACT
Long non-coding RNAs (LncRNAs) have been recently regarded as systemic regulators in multiple biologic processes including tumorigenesis. In this study, we observed the expression of lncRNA lnc-sox5 was significantly increased in colorectal cancer (CRC). Despite the CRC cell growth, cell cycle and cell apoptosis was not affected by lnc-sox5 knock-down, lnc-sox5 knock-down suppressed CRC cell migration and invasion. In addition, xenograft animal model suggested that lnc-sox5 knock-down significantly suppressed the CRC tumorigenesis. Our results also showed that the expression of indoleamine 2,3-dioxygenase 1 (IDO1) was significantly reduced by lnc-sox5 knock-down and therefore modulated the infiltration and cytotoxicity of CD3+CD8+T cells. Taken together, these results suggested that lnc-sox5 unbalances tumor microenvironment to regulate colorectal cancer progression.
Introduction
Colorectal cancer (CRC) is the third most common cancer and ranked the fourth lethal tumor among all human cancers worldwide.Citation1,2 The combined use of irinotecan, oxaliplatin and oral form of 5-fluorouracil (5-FU) has shown significant therapeutic efficacy in human CRC.The treatment of radiotherapy with/without chemotherapy and radical surgery has become the gold standard treatment of CRC. However, the toxicity of radiotherapy and chemotherapy had been reported to be associated with morbidity and mortality of CRC.Citation3-5 Therefore, identifying novel biomarkers for CRC diagnosis and therapy is urgently needed.
Long non-coding RNAs (LncRNAs) are generally defined as untranslational transcripts composed of more than 200 nucleotides in length and,Citation6-8 and associated with many important cellular processes and pathogenesis. Numerous lncRNAs are involved in tongue carcinogenesis and metastasis. HOX transcript antisense RNA (HOTAIR), which is the first identified lncRNA that plays a critical oncogenic role through epigenetic regulatory mechanisms, can directly stimulate chromatin modifications related to tumor invasiveness and metastasis.Citation9,10 It has been reported that H19 plays as a tumor suppressor in CRC by directly involved in the activation of the downstream gene expression of c-Myc.Citation11 The molecular mechanisms underlying the altered expression of PCAT-1 in CRC remain unclear.Citation12 It has been reported HULC is ultrahighly expressed in hepatic colorectal metastasis.Citation12 Despite many lncRNAs expressed in CRC, the role and the mechanism of them in tumorigenesis don't clearly identified.
In this study, we observed lnc-sox5 was increased in CRC. We notified that lnc-sox5 knock-down did not affect the growth of colon cancer cells in vitro and in immune deficient animals, but significantly suppressed tumorigenesis in immune proficient animals. Extensively, lnc-sox5 knock-down dramatically promoted the infiltration and the cytotoxicity of CD3+CD8+ CTLs. We observed lnc-sox5 knock-down suppressed IDO1 expression to unbalance the tumor microenvironment. These results suggest lnc-sox5 may contribute to the diagnosis and therapeutic target for CRC.
Materials and methods
Clinical tissues
Total 52 patients that have never received therapy before surgery were recruited from First Affiliated Hospital of Sun Yat-Sen University in this study. Tissues were collected after acquirement of informed consent from patients. CRC tissues and paired adjacent normal tissues were collected and rapid frozen in liquid nitrogen for subsequent q-RT-PCR analysis. All protocols have been approved by Ethics Committee of First Affiliated Hospital of Sun Yat-Sen University.
Cell culture
Human colorectal cancer cell line Caco-2 and mouse colorectal cancer cell line MC-38 were obtained from First Affiliated Hospital of Sun Yat-Sen University and cultured in RPMI 1640 (Hyclone) with supplement of 10% FBS (Gibco-BRL; Invitrogen) at a humidity of 5% CO2 at 37 °C.
MTT
CRC cell proliferation was evaluated by MTT ([3-(4,5-dimethylthiazol-2-yl)-2,5 -diphenyltetrazolium bromide) assay. In brief, 5.0 × 103 cell/well cells were seeded in triplicate in 96-well plates and cultured for 24 h. Cells were transfected with siRNAs for 24 h for subsequent MTT assay with MTT cell proliferation assay kit (Abnova, Walnut, CA), according to manufacturer's protocols. Absorbance of each well was measured at 570 nm with a spectrophotometer (ELISA reader, Dynatech Laboratories, Chantilly, VA). The sequence of these siRNAs were listed at .
Table 1. The sequences of primers and siRNAs used in this study.
Transwell invasion assay
Caco-2 and MC-38 cell invasion capacity were assessed using Trans-well Chamber Cell Culture (10-μm pore membrane, BD Biosciences). 1 * 105 cells which were transfected with 100nM siRNA in 100 μl of serum-free medium were added to the top chamber of 24-well plates that has been pre-added with Matrigel. The bottom well contained growth medium with 20% FBS. Trans-well chambers were placed at 37 °C for 36 h. Cells in chamber were fixed with methanol for 30 min and then staining with Giemsa for 15–30 min. Invaded cells were finally observed under a microscope.
Wound scratch assay
A total of 5 × 105 cells, which were transfected with lnc-sox5 siRNA, were seeded in 6-well culture plates and grown to 85–90% confluence. Scratch was then vertically created by a p200 pipet tip in the cell monolayer. After a period of 24 h incubation in normal condition, images for cell growth around wounding were captured. Number of cell number around wounding calculated and was normalized to Mock.
Western blot
Lysates from tumor samples, Caco-2 cells and MC-38 cells were prepared with RIPA Lysis buffer (Beyotime, China) containing protease inhibitor cocktail. Protein samples were loaded for sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. After blocking with 5% fat-free milk, the membrane was probed with primary anti-IDO1 antibody (dilution 1:1000; Santa Cruz Biotechnology), anti-E-cadherin antibody (dilution 1:2000; Santa Cruz Biotechnology), anti-N-cadherin antibody (dilution 1:2000; Santa Cruz Biotechnology), anti-Vimentin antibody (dilution 1:2000; Santa Cruz Biotechnology) anti-β-actin (dilution 1:2000; Santa Cruz Biotechnology) antibody. After washing, the membrane was incubated with horseradish peroxidase-conjugated (HRP) secondary antibody for 1 hour. The signal was visualized using the ECL detection system (Thermo Fisher, USA) and quantified by densitometry using Quantity One software (Bio-Rad, Hercules, CA, USA).
qRT-PCR
Total RNA were purified from CRC tissues or colon cancer cells as manufacturer's instructions described. To detect the expression of lnc-sox5, cDNA were synthesized by Reverse Transcription Kit (Takara, Dalian, China). qRT-PCR was performed using SYBR Green Master Mix (Roche, Basel, Switzerland). All primers were listed at . Relative mRNA levels were determined by the comparative Ct method after normalization to β-actin.
Animal model
Male C57BL/6 mice and Balb/c nude mice with 7-week-age were obtained from SLAC Laboratory Animals Co Ltd (Shanghai, China). Animals protocols have been approved by animal Ethics Committee of First Affiliated Hospital of Sun Yat-Sen Univesity and performed according to the institutional guidelines. Subcutaneous tumor transplantation was performed by 2.0*106 of Caco-2 cells or MC-38 cells injection in the right flank of mice. Tumor was removed after 4 weeks transplantation and the T cell phenotype was analyzed by flow cytometry.
Flow cytometry
Flow cytometry was performed with Canto II (BD Bioscience). Fluoro-chrome conjugated anti-CD3 (145–2C11, eBioscience), anti-CD4 (RM4–5, eBioscience), anti-CD8 (eBioH35–17.2, eBioscience), anti-Foxp3 (FJK-16s, eBioscience) and anti-Granzyme B (NGZB, eBioscience). Granzyme B staining was performed with intracellular staining kit from BD Bioscience. While Foxp3 staining was done with nuclear protein staining kit from eBioscience (San Diego, CA). FACS data were analyzed using FlowJo software (Tree Star, Ashland, OR).
L-kynurenine determination
Caco-2 cell or MC-38 cells, which were transfected with mlnc-sox5 or hlnc-sox5 siRNAs, were seeded into 24 well plate at 2.0*10ˆ5 cell/well. Measurements of IDO1 functional activity in terms of L-kynurenine production in plasma or cell supernatants were as described.Citation13,14
Statistical analysis
Statistical analysis was performed for all experiments using Student's t-test. Values of p inferior to 0.05 were considered significant. All tests were done using Prism 6 software (Graph Pad Prism6).
Results
lnc-sox5 expression is increased in human CRC
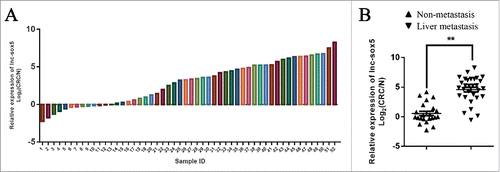
To identify aberrantly expressing LncRNAs in CRC, clinic tumor samples were examined (Fig. S1). The q-PCR analysis showed that lncRNA lnc-sox5 was significantly increased lncRNA in clinic colorectal cancer samples (). To determine the role of lnc-sox5, several colorectal cancer cell lines were subject to q-PCR analysis. The results revealed that Caco-2 cells showed the highest lnc-sox5 expression (Fig. S2). To confirm this data, 52 pairs of clinic tumor samples were subject to qRT-PCR for testing the level of lnc-sox5 (). In addition, the q-PCR analysis also revealed that lnc-sox5 level was significantly increased in liver metastatic CRC (). These results suggested that increased lnc-sox5 might regulate the progression of CRC.
Figure 1. lnc-sox5 expression in clinic CRC samples. (A) The relative expression of lnc-sox5 in 52 pairs of CRC samples was determined by qRT-PCR. (B) The relative expression of lnc-sox5 in metastatic and non-metastatic CRC samples was determined by qRT-PCR. All results were normalized by β-actin. Fold means log2(C/N), C represents CRC, N represents matched adjacent normal tissue. All results were shown as mean ± SEM * means p < 0.01, ** means p < 0.01 and *** means p < 0.001.
lnc-sox5 inhibition suppresses CRC cell invasion and migration
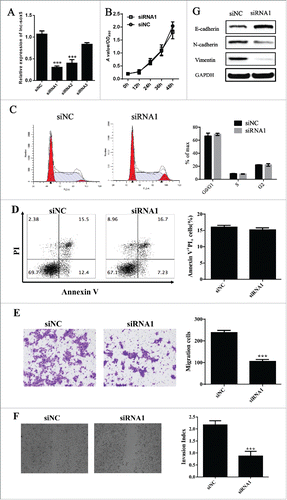
To address the physiologic effect of lnc-sox5 on CRC, we first suppressed the expression of lnc-sox5 by small interference RNA (siRNA, ). However, Caco-2 cell growth (), cell apoptosis () and cell cycle () did not affected by lnc-sox5 inhibition in vitro. Interestingly, both invasion () and migration () were significantly suppressed by lnc-sox5 knock-down. In addition, EMT related genes were all decreased by lnc-sox5 knock-down (). These results suggested that the aberrant lnc-sox5 expression modulated CRC metastasis.
Figure 2. lnc-sox5 knock-down suppressed Caco-2 cells migration and invasion. (A) The efficacy of siRNAs targeting to lnc-sox5 was examined by qRT-PCR. (B) MTT assays were performed for testing the effect of lnc-sox5 on Caco-2 cell growth. Flow cytometry was used to assess the effect of lnc-sox5 on Caco-2 cell cycle (C) and cell apoptosis (D). (E) The invasion of Caco-2 cells was examined by trans-well assays. (F) Wound scratch assay was used to determine the effect of lnc-sox5 on Caco-2 cell migration. (G) EMT related molecules in Caco-2 cells which were lnc-sox5 knock-down were detected by western blot. qRT-PCR results were normalized by β-actin. All results were shown as mean ± SEM * means p < 0.01, ** means p < 0.01 and *** means p < 0.001.
lnc-sox5 inhibition impairs CRC tumor microenvironment
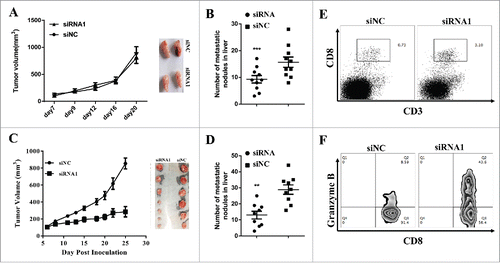
To confirm the role of lnc-sox5 in vivo, we first construct a siRNA1 stably expressing Caco-2 cells. Xenograft nude mouse model showed that lnc-sox5 knock-down did not affect the tumor growth in vitro () but the liver metastasis was significantly suppressed (). We also constructed a mouse lnc-sox5 knock-down colon cancer cell line (MC-38). Consistent to Caco-2 cells, the cell cycle and the apoptosis in MC-38 cell were not suppressed by mlnc-sox5 knock-down (Fig. S3). However, both tumor growth () and liver metastasis () of MC-38 cell in immune proficient mouse were significantly suppressed by lnc-sox5 knock-down. To investigate why lnc-sox5 knock-down suppressed tumor growth in immune proficient mice, the tumor microenvironment were subject to flow cytometry analysis. The frequency of CD3+CD8+CTLs was dramatically increased in lnc-sox5 knock-down tumors (). Furthermore, the cytotoxicity of these CD3+CD8+CTLs which was always marked as Granzyme B+ was also enhanced by lnc-sox5 knock-down (). These results suggested that lnc-sox5 might intrinsically promoted colon cancer cell metastasis and indirectly unbalanced tumor microenvironment to suppress colon cancer progression.
Figure 3. lnc-sox5 regulates the tumor growth and tumor microenvironment in immune proficient mice. (A) Balb/c nude mice were inoculated with 2.0*10ˆ6 Caco-2 cells which were siRNA1 or siNC stably expressing and the tumor volume was measured. (B) The metastatic nodules in liver was counted in hlnc-sox5 knock-down or NC groups. (C) Immune proficient mice were inoculated with 2.0*10ˆ6 MC-38 cells which were siRNA or siNC stably expressing and the growth curve was measured. (D) The metastatic nodules in liver was counted in mlnc-sox5 knock-down or NC groups. (E) The frequency of CD3+CD8+ CTLs was determined by flow cytometry. (F) The cytotoxicity of CD8+CTLs was assessed by intracellular staining with Granzyme B antibody and subsequently detected by flow cytometry. All results were shown as mean ± SEM * means p < 0.01, ** means p < 0.01 and *** means p < 0.001.
lnc-sox5 suppresses IDO1 expression to impair CRC tumor microenvironment
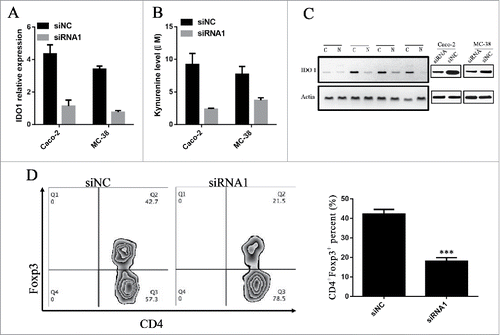
To investigate how lnc-sox5 impairs colon tumor microenvironment, the indoleamine-2,3-dioxygenase 1 (IDO1) level was determined by q-PCR in Caco-2 cells and MC-38 cells after lnc-sox5 knockdown (). The q-PCR analysis revealed that lnc-sox5 knockdown significantly suppressed the mRNA level of IDO1. The IDO1 protein level was also confirmed in MC-38 cells and Caco-2 cells and CRC samples (). To confirm these results, the L-kynurenine (KYN) level in the supernatant of Caco-2 and MC-38 cells, which were lnc-sox5 knock-down, were also determined. Consistently, lnc-sox5 knock-down significantly suppressed KYN production (). It seems that IDO1 was the direct target of lnc-sox5 in CRC cells. To address this question, RNA immune-precipitation (RIP) assays were performed. However, RIP analysis revealed that lnc-sox5 could not directly binding to IDO1 (Fig. S4). IDO1 has been reported to promote the differentiation of Tregs in tumor microenvironment. As expected, flow cytometry analysis suggested that lnc-sox5 knock-down remarkably suppressed Treg frequency (). These results demonstrated that lnc-sox5 might indirectly regulate IOD1 level to unbalance the tumor microenvironment.
Figure 4. lnc-sox5 regulates the level of IDO1 to unbalance colon tumor microenvironment. (A) The level of IDO1 in Caco-2 and MC-38 cells was detected by q-RT-PCR. (B) The L-kynurenine (KYN) level in the supernatant of Caco-2 and MC-38 cells was examined. (C) The protein level of IDO1 in clinical CRC (C) or Normal (N) tissues and siRNA transfected MC-38 cells were examined by western blot. (D) The frequency of CD4+Foxp3+ Tregs in immune proficient mice which were inoculated with MC-38 cells was tested by flow cytometry. All results were shown as mean ± SEM * means p < 0.01, ** means p < 0.01 and *** means p < 0.001.
Discussion
Many LncRNAs aberrantly expressed and acted as regulators in human tumorigenesis. These LncRNAs are epigenetic regulation of protein coding genes. Recently, multiple LncRNAs have been reported to play vital role in colorectal carcinogenesis including H19,Citation15-17 CCAT1Citation18-21 and CCAT2,Citation22 etc. In this study, we identified that lnc-sox5 was correlated with colorectal cancer progression. The aberrant expression of lnc-sox5 in CRC was subsequently verified to intrinsically suppress the metastasis of CRC and also to modulate the CD8+T cell function.
Today there is a great debate about the role of inflammation in control and regulation of tumor microenvironment directly influencing the development of neoplasms. Inflammation plays a fundamental role in tumor dynamic and is accepted as one of the hallmarks of cancer.Citation23,24 Currently there are 2 marked ways by which inflammation is associated with cancer development, an intrinsic pathway characterized by the gene expression and an extrinsic pathway, characterized by the formation of an inflammatory microenvironment.Citation25 In this study, we identified that lnc-sox5 played as a tumor oncogene to regulate the progression of CRC. It has been reported IDO1 promotes the differentiation and infiltration of Tregs in tumor microenvironment and therefore regulates tumor progression.Citation26 In this study, IDO1 expression was remarkably reduced by lnc-sox5 knock-down in colon cancer cells. The cytotoxicity of CD8+CTLs is suppressed by Tregs by few well-known signals including inhibitory immume checkpoint molecules, inducing CTL exhaustion and inducing CTL apoptosis by Fas/FasL pathway.Citation27 Taken together, increased lnc-sox5 promotes the expression of IDO1 in CRC cells and subsequently promotes the infiltration or differentiation of Treg cells in tumor microenvironment.
In this study, to investigate the potential role of lnc-sox5 in the progression of colorectal cancer, the expression of lnc-sox5 in 52 pairs of CRC samples and adjacent normal colon clinic samples were determined by qRT-PCR analysis. The results showed that lnc-sox5 was significantly increased in CRC tumor tissues. Extensive experiments revealed that lnc-sox5 modulated the CD8+T cells infiltration by regulating IDO1 expression and therefore suppressed the tumorigenesis of CRC. All these results suggest lnc-sox5 may contribute to the diagnosis and therapeutic target for CRC.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
Conceived and supervised the study by Liang Wang from Gastrointestinal Surgery Center, 1st Affiliated Hospital of Sun Yat-Sen University. The cell lines are constructed by Zhenxian Zhao from Department of Pancreato-Biliary Surgery, 1st Affiliated Hospital of Sun Yat-Sen University. The construction of animal model, flow cytometry analysis and the manuscript writing is performed by Kaiming Wu from Gastrointestinal Surgery Center, 1st Affiliated Hospital of Sun Yat-Sen University. We also thank Kuanzhi Liu for the animal grooming, Jian Zhang for the Western blot assay, Guanghua Li for the cell culture and manuscript checking.
Supplemental_Material.pdf
Download PDF (144.5 KB)Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Fearon ER. Molecular genetics of colorectal cancer. Ann Rev Pathol 2011; 6:479-507; PMID:21090969; https://doi.org/10.1146/annurev-pathol-011110-130235
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65:87-108
- Breugom AJ, van Gijn W, Muller EW, Berglund A, van den Broek CB, Fokstuen T, Gelderblom H, Kapiteijn E, Leer JW, Marijnen CA, et al. Adjuvant chemotherapy for rectal cancer patients treated with preoperative (chemo)radiotherapy and total mesorectal excision: a Dutch Colorectal Cancer Group (DCCG) randomized phase III trial. Ann Oncol 2015; 26:696-701; https://doi.org/10.1093/annonc/mdu560
- Deng Q, Huang CM, Chen N, Li L, Wang XD, Zhang W, Bi F, Tang QL, Li ZP, Wang W. Chemotherapy and radiotherapy downregulate the activity and expression of DNA methyltransferase and enhance Bcl-2/E1B-19-kDa interacting protein-3-induced apoptosis in human colorectal cancer cells. Chemotherapy 2012; 58:445-53; PMID:23364257; https://doi.org/10.1159/000345916
- Kimura K, Mishima H, Ohashi N, Itoh N, Arikawa T, Kiyota Y, Nagata H, Suzumura K, Miyachi M, Mizumatsu S, et al. [A case of colorectal cancer with pelvic recurrence treated by systemic chemotherapy and radiotherapy]. Gan To Kagaku Ryoho Cancer Chemother 2014; 41:1719-21
- Huarte M. RNA. A lncRNA links genomic variation with celiac disease. Science 2016; 352:43-4; PMID:27034364; https://doi.org/10.1126/science.aaf6015
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013; 500:598-602; PMID:23945587; https://doi.org/10.1038/nature12451
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013; 154:26-46; PMID:23827673; https://doi.org/10.1016/j.cell.2013.06.020
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464:1071-6; PMID:20393566; https://doi.org/10.1038/nature08975
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011; 71:6320-6; PMID:21862635; https://doi.org/10.1158/0008-5472.CAN-11-1021
- Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 2006; 66:5330-7; PMID:16707459; https://doi.org/10.1158/0008-5472.CAN-06-0037
- Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol 2013; 30:588; PMID:23640607; https://doi.org/10.1007/s12032-013-0588-6
- Curti A, Pandolfi S, Valzasina B, Aluigi M, Isidori A, Ferri E, Salvestrini V, Bonanno G, Rutella S, Durelli I, et al. Modulation of tryptophan catabolism by human leukemic cells results in the conversion of CD25- into CD25+ T regulatory cells. Blood 2007; 109:2871-7; PMID:17164341
- Fallarino F, Grohmann U, Hwang KW, Orabona C, Vacca C, Bianchi R, Belladonna ML, Fioretti MC, Alegre ML, Puccetti P. Modulation of tryptophan catabolism by regulatory T cells. Nat Immunol 2003; 4:1206-12; PMID:14578884; https://doi.org/10.1038/ni1003
- Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res 2002; 62:6442-6; PMID:12438232
- Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang J, Dong N, He J, Sun Q, Lv G, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget 2016; 7:22159-73; PMID:26989025
- Nakagawa H, Chadwick RB, Peltomaki P, Plass C, Nakamura Y, de La Chapelle A. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc Natl Acad Sci U S A 2001; 98:591-6; PMID:11120891; https://doi.org/10.1073/pnas.98.2.591
- McCleland ML, Mesh K, Lorenzana E, Chopra VS, Segal E, Watanabe C, Haley B, Mayba O, Yaylaoglu M, Gnad F, et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Invest 2016; 126:639-52; PMID:26752646; https://doi.org/10.1172/JCI83265
- Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 2014; 24:513-31; PMID:24662484; https://doi.org/10.1038/cr.2014.35
- Ye Z, Zhou M, Tian B, Wu B, Li J. Expression of lncRNA-CCAT1, E-cadherin and N-cadherin in colorectal cancer and its clinical significance. Int J Clin Exp Med 2015; 8:3707-15; PMID:26064266
- Zhao W, Song M, Zhang J, Kuerban M, Wang H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int J Clin Exp Pathol 2015; 8:14131-40; PMID:26823726
- Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 2013; 23:1446-61; PMID:23796952; https://doi.org/10.1101/gr.152942.112
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57-70; PMID:10647931; https://doi.org/10.1016/S0092-8674(00)81683-9
- Mantovani A. Cancer: Inflaming metastasis. Nature 2009; 457:36-7; PMID:19122629
- Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol 2010; 22:231-7; PMID:20144856; https://doi.org/10.1016/j.coi.2010.01.009
- Bonanno G, Mariotti A, Procoli A, Folgiero V, Natale D, De Rosa L, Majolino I, Novarese L, Rocci A, Gambella M, et al. Indoleamine 2,3-dioxygenase 1 (IDO1) activity correlates with immune system abnormalities in multiple myeloma. J Transl Med 2012; 10:247; PMID:23232072; https://doi.org/10.1186/1479-5876-10-247
- Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013; 14:1014-22; PMID:24048123; https://doi.org/10.1038/ni.2703
