ABSTRACT
Defects in the DNA damage response (DDR) are associated with multiple diseases, including cancers and neurodegenerative disorders. Emerging evidence indicates involvement of RNA-binding proteins (RBPs) in DDR. However, functions of RBPs in the DDR pathway remain elusive. We have shown previously that the RNA-binding protein RBM14 is required for non-homologous end joining (NHEJ). Here we show that RBM14 is required for efficient recruitment of XRCC4 and XLF to chromatin and the release of KU proteins from chromatin upon DNA damage. Failure of this process leads to accumulation of double-strand breaks (DSBs) in cells. Thus RBM14 plays crucial role in regulation of NHEJ upon DNA damage.
KEYWORDS:
DSBs occur as a result of replication errors and DNA damaging agents. Cells have elaborate mechanisms to respond to DNA damage. Networks of DDR pathways are coordinated to detect and repair DNA damage, regulate cell cycle and transcription, and determine the cell fate. One of the main pathways to repair DSBs in mammalian cells is NHEJ. DNA-PK is a crucial component of both NHEJ machinery and the V(D)J recombination apparatus. It is a PI-3-like kinase, and is activated upon association with DNA. Biochemical and genetic data have revealed DNA-PKcs to be a component of a large catalytic subunit of DNA-PK that contains a regulatory factor termed KU.Citation1-Citation6 The DNA-end sensor KU70/80 heterodimer together recognizes DSB ends and recruits DNA-PKcs to initiate the NHEJ process. Then, the DSB ends are ligated by the DNA ligaseIV complex, which includes the X-ray cross-complementing gene 4 (XRCC4) and XRCC4-like factor (XLF).Citation7 DNA ligase IV is stabilized by the scaffold proteins XRCC4 and XLF, which form long helical filaments.Citation8
Emerging evidence suggests that RBPs physically interact with DNA lesions and thus are involved in DNA damage response.Citation9-Citation14 RBM14 contains RNA recognition motifs (RRMs) and an unstructured domain called the prion-like domain (PLD). We identified RBM14 as a gene that radio-sensitizes GBM (glioblastoma multiforme) spheres when knocked down by a human genome-wide shRNA screen. RBM14 interacts with KU, and knockdown of RBM14 inhibits NHEJ significantly. Importantly, knockdown of RBM14 reduces tumorigenicity and also radio-sensitizes GBM stem-like cells in vivo.Citation15 RBM14 is highly expressed in embryonic tissues and in stem cells, and it has been thought that RBM14 controls transcription-coupled alternative splicing in a manner that depends on the promoter.Citation16,Citation17 RBM14 is an essential paraspeckle component, and mediates a key interaction linking several other essential proteins into the network.Citation18 Paraspeckles are a nuclear body which consists of nuclear RNA-binding proteins, including RBM14, FUS, TAF15, and EWSR1, and the long noncoding RNA NEAT1 (Nuclear Paraspeckle Assembly 1/MENϵ/β). Paraspeckles are induced by stress, such as viral infection, proteasome inhibition, and differentiation,Citation19-Citation22 however, the function of this nuclear body is not well understood.
Interestingly, the other paraspeckle proteins, such as FUS and NONO, have also been implicated in DDRs. NONO is recruited to damaged DNA sites, and stimulates NHEJ and represses HR.Citation12 FUS is also recruited to damaged DNA sites, however, knockdown of FUS inhibits both NHEJ and HR.Citation11,Citation23-Citation26 It has been shown that FUS directly interacts with histone deacetylase 1 (HDAC1), and that this interaction is required for the function of FUS in DDR. Knockdown of FUS inhibits γH2AX formation in response to DSBs,Citation23 whereas RBM14 knockdown induces prolonged γH2AX foci.Citation15 How these RBPs are involved in DNA damage response including DNA repair remains elusive.
Despite the considerable number of RBPs that have been implicated in the DNA damage response, little is known about their roles in DSB repair pathways. To gain new insights into DSB repair mechanisms, we sought to characterize functions of RBM14 in DSB repair.
Results and discussion
We have shown previously that RBM14 interacts with KU, and that knockdown of RBM14 inhibits NHEJ in undifferentiated GBM stem-like cells significantly.Citation15 We tested whether RBM14 is also required for NHEJ in non-stem cells. Knockdown of RBM14 sensitizes U2OS as well as 293T cells to IR (Fig. S1B and data not shown). As in the case with GBM spheres, knockdown of RBM14 inhibited NHEJ significantly (Fig. S2B), indicating that RBM14s function in NHEJ is not limited to undifferentiated cells. The other main pathway to repair DSBs is homologous recombination (HR). Thus, we tested the effects of RBM14 knockdown on the efficiency of HR by the GFP-based HR reporter system.Citation27 BRCA1 knockdown showed inhibition of HR, whereas the HR efficiency was not significantly affected by RBM14 knockdown (Fig. S2A), suggesting that RBM14 specifically controls the NHEJ pathway.
Figure 1. Proper association and dissociation of NHEJ factors depend on RBM14. (A) RBM14 is required for recruitment of XRCC4 and XLF to damaged chromatin. U2OS cells were treated with 500 µM phleomycin for the indicated times. Cells were fractionated in lysis buffer containing RNaseA (chromatin; left) or lysed as whole cell extracts (right). (B) RBM14 knockdown inhibits release of KU proteins from damaged chromatin. U2OS cells were treated with 500 µM phleomycin for 60 min, and then left to recover following phleomycin removal. Cells were collected at the indicated time points. KD: knockdown.
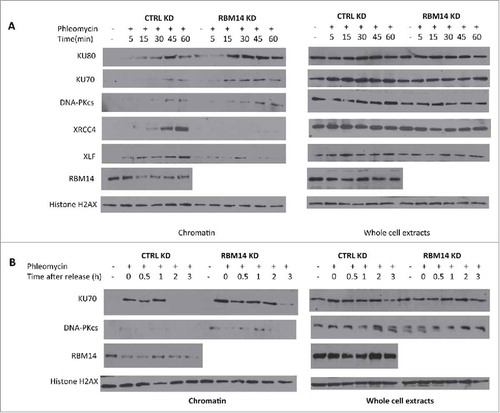
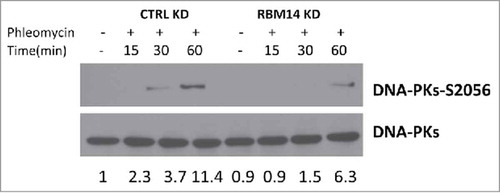
Figure 2. DNA-PK autophosphorylation is compromised in RBM14 knockdown cells. Immunoblotting of whole cell extracts from U2OS cells transfected with the indicated siRNA. Cells were irradiated (3 Gy), and collected at the indicated time points after irradiation. KD: knockdown.
To further explore how RBM14 contributes to NHEJ, we examined the effects of RBM14 knockdown on KU recruitment and dissociation by monitoring accumulation of KU proteins in RNase A-resistant chromatin fractions after treating cells with phleomycin. This procedure is based on successive detergent extractions following treatment of cells with a DNA damaging agent. The final fractions contain proteins that are tightly bound to chromatinCitation13 (hereafter chromatin fraction). NHEJ factors including, KU, XRCC4, and XLF were accumulated in the chromatin fractions upon phleomycin treatment in control cells. We observed significantly more chromatin-bound KU proteins in RBM14 knockdown cells although the total protein levels of NHEJ proteins remained the same as in the control knockdown cells (). DNA-PKcs proteins were hardly detectable in the chromatin fractions in control knockdown cells, which might be because of transient recruitment of DNA-PKcs. However, we observed an increased amount of DNA-PKcs protein in the chromatin fractions from RBM14 knockdown cells (). Strikingly, RBM14 knockdown abolished accumulation of DNA ligase IV components, XRCC4 and XLF, in chromatin fraction (), indicating that persistent KU proteins at the DSB sites might inhibit recruitment of the ligase IV complex to damaged chromatin in RBM14 knockdown cells. To test this, we monitored KU protein levels in RNase A-resistant chromatin fractions after releasing cells from the phleomycin treatment. In control cells, KU protein was detected in the RNaseA-resistant chromatin fractions up to 1 hour, but not detectable at 2 hours time points after releasing from the phleomycin treatment. In contrast, KU persisted in the chromatin fraction at 3 hours post-release in RBM14 knockdown cells. Therefore, RBM14 knockdown caused KU80 and KU70 persistent binding to chromatin after treating cells with a pulse of phleomycin ().
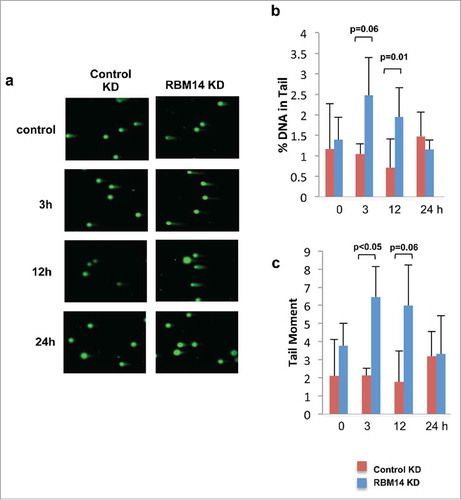
We have shown previously that DNA-PKcs autophosphorylation at Ser2056 was compromised in RBM14-knockdown GBM spheres.Citation15 The same effect was detected with irradiated 293T cells (). The classic model indicates that the NHEJ process occurs in a step-wise manner; (1) recruitment of KU heterodimer to DSB ends, (2) DNA-PKcs binding to KU proteins in DNA-dependent manner, (3) autophosphorylation of DNA-PKcs, which induces conformational changes of DNA-PKcs, (4) recruitment of the DNA ligase IV complex, (5) DNA-end ligation.Citation28,Citation29 However, recent studies indicate that KU-mediated DSB repair might take place by a dynamic multi-protein complex. Another intrinsically disordered protein, APLF, has been shown to act as a scaffold for the KU-DNA-PKcs complex and the DNA-ligase IV/XRCC4 complex by biochemical reconstitution of DSB-end ligation with purified proteins.Citation30 In this model, dissociation of DNA-PKcs is not required, but instead APLF provides space for the DNA-ligase IV complex at DSB sites while DNA-PKcs is still interacting with KU proteins. A non-catalytic function of the DNA-ligase IV complex has also been shown. DNA ligase IV was shown to be required for stimulation of DNA-PKcs autophosphorylation.Citation31 Furthermore, 2-stage synapsis of DNA ends has been detected by using single-molecule imaging with X.laevis egg extracts. DNA ends are initially tethered in a long-range complex whose formation requires KU proteins and DNA-PKcs. The ends are closely aligned by XLF, the non-catalytic function of Ligase IV/XRCC4, and DNA-PK activity.Citation32 Our results from the cell-free chromatin fractionation assay indicate that the RNA-binding protein RBM14 is required for the efficient recruitment of DNA-ligase IV complex to DNA damage sites. It is possible that RBM14 also acts as a scaffold protein to link between KU-DNA-PKcs and DNA-ligase IV/XRCC4 complexes as in the case of APLF. Autophosphorylation of DNA-PKcs is compromised in RBM14 knockdown cells. This could be caused by inefficient recruitment of DNA-ligase IV complex. We also observed accumulation of KU proteins in damaged chromatin fractions, indicating that KU dissociation/degradation depends on DNA-ligase IV recruitment and autophosphoryltion of DNA-PKcs that require RBM14. Consistent with these results, persistent DSBs were observed in RBM14 knockdown cells. DSBs were repaired within 3 hours after irradiation in control cells, whereas RBM14 knockdown cells exhibited comet tails even at 12 hours after irradiation, indicating that DSB ends were not ligated (). It is not known whether RBM14 recruits the DNA-ligase IV complex directly through physical interaction. We did not observe interactions between RBM14 and the DNA-ligase IV complex components XRCC4 and XLF, while KU protein was immunoprecipitated by anti-RBM14 antibody (Fig. S3). However, this does not exclude weak and/or transient interactions.
Figure 3. RBM14 knockdown cells are not able to repair DNA damage efficiently. (A) Presence of DNA damage at sequential time points after 3Gy of IR was accessed by single-cell gel-electrophoresis assay under neutral conditions (neutral comet assay). (B) Quantification of the percentages of cells with comet tails at the indicated time points. (C) Quantification of the tail moments at the indicated time points. KD: knockdown.
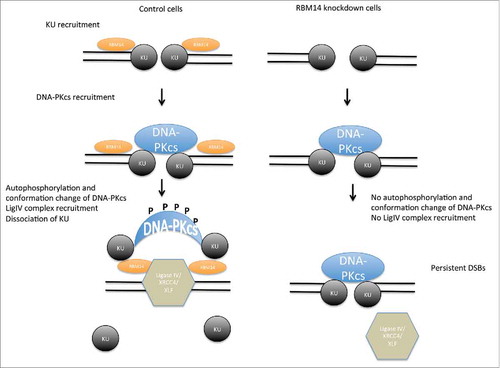
In conclusion, we demonstrated that the RNA-binding protein RBM14 plays a crucial role in coordinating recruitment and dissociation of NHEJ proteins, and that failure of this process leads to accumulation of DSBs in cells (, model).
Figure 4. Model of NHEJ factor regulation. The KU heterodimers are recruited to DSB sites, and then DNA-PKcs complex is recruited in KU-dependent manner, securing the DSB end synapsis. RBM14 is required for the recruitment of DNA-ligase IV complex and autophosphorylation of DNA-PKcs which causes conformation change of DNA-PKcs. In the absence of RBM14, the DNA-ligase IV complex is not recruited to DSB sites. KU proteins persist on damaged chromatin, and ligation does not occur.
Methods
Plasmids, SiRNAs and antibodies
CoAA-Flag was a gift from Lan Ko. Phleomycin was acquired from Sigma (P9564). siRNAs RBM14–6 (SI02637229), RBM14–7 (SI02637236), 53BP1–8 (SI00079961), BRCA1–14 (SI02664361), and negative control siRNA (Allstar Negative control, 1027281) were obtained from Qiagen. All siRNAs were transfected with lipofectamine RNAiMAX from Thermofisher (13778). Antibodies used in western blotting were RBM14 (1:1000, abcam ab70636), Ku70 (1:250, abcam ab3114), Ku80 (1:250, Fisher Scientific MS285P0), XRCC4 (gift from Dr. Patrick Calsou), XLF (1:250, abcam ab33499) H2AX (1:10,000, abcam ab11175).
Cell culture
U2OS and 293T cells were grown in DMEM with glutamine and 4.5 g/L glucose (Corning Cellgro 10–013-CV) supplemented with 10% Fetal Bovine Serum (Sigma F2442) in 5% CO2 incubator at 37˚C.
Biochemical fractionation
Cells were fractionated essentially as in,Citation13 with minor modifications. After treatment with phleomycin, cells were washed with PBS and harvested by scraping into 250 microliters of lysis buffer (50-mM Hepes pH 7.5, 150-mM NaCl, 1-mM EDTA) containing 0.05% NP-40 supplemented with 5U/mL RNAse A. 50μL were removed to assess total protein levels, and the remainder was incubated on ice for 5 min then centrifuged for 5 min at 1000 g. The supernatant was removed (Fraction I), and the pellet was resuspended in 200 μL of the same buffer and centrifuged as above. This supernatant (Fraction II) was removed and the pellets were resuspended in lysis buffer containing 0.5% NP-40. Samples were incubated on ice for 45 minutes, then centrifuged at 16,000 g for 15 minutes. The final supernatant was removed (Fraction III) and the pellets were resuspended in lysis buffer containing 1% Triton, sonicated, then analyzed via western blot (Chromatin Fraction).
HR GFP assay
HR GFP assays were performed as described in.Citation27 Cells were reverse transfected with siRNAs and I-SceI, analyzed with a FACscaliber (BD Biosciences) and quantified in FlowJo.
NHEJ GFP assay
NHEJ efficiency was analyzed with a plasmid based assay described in,Citation30 Cells were co-transfected with a Hind-III digested reporter plasmid and DS-red transfection control. Double positive cells were counted with a FACscaliber (BD biosciences) and repair efficiencies quantified in FlowJo
Clonogenic assay
Clonogenic analysis was performed as described previously.Citation15 U2OS cells were plated at described densities and irradiated the same day at indicated doses in a CIXD Xtrahl X-ray irradiator. Colonies were allowed to form for 14 d before fixing and staining with 0.5% crystal violet in 50% methanol.
Neutral comet assay
Cells were treated with or without IR at different time points, then cells were collected and analyzed by single cell gel electrophoresis assay (TREVIGEN) under neutral condition according to manufacturer's protocol. 100 cells of each group were quantified by using CometScore.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1317419_Supplemental_Material.zip
Download Zip (1.2 MB)Acknowledgments
We thank Dr. Jeremy Stark (the City of Hope) for providing the GFP-based HR assay system. We thank Dr. Vera Gorbunova (University of Rochester) for providing the plasmid-based NHEJ assay system.
References
- Zha S, Jiang W, Fujiwara Y, Patel H, Goff PH, Brush JW, Dubois RL, Alt FW. Ataxia telangiectasia-mutated protein and DNA-dependent protein kinase have complementary V(D)J recombination functions. Proc Natl Acad Sci U S A 2011; 108:2028-33; PMID:21245310; https://doi.org/10.1073/pnas.1019293108
- Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell 2009; 34:285-97; PMID:19450527; https://doi.org/10.1016/j.molcel.2009.04.025
- Gapud EJ, Dorsett Y, Yin B, Callen E, Bredemeyer A, Mahowald GK, Omi KQ, Walker LM, Bednarski JJ, McKinnon PJ, et al. Ataxia telangiectasia mutated (Atm) and DNA-PKcs kinases have overlapping activities during chromosomal signal joint formation. Proc Natl Acad Sci U S A 2011; 108:2022-7; PMID:21245316; https://doi.org/10.1073/pnas.1013295108
- Gapud EJ, Sleckman BP. Unique and redundant functions of ATM and DNA-PKcs during V(D)J recombination. Cell Cycle 2011; 10:1928-35; https://doi.org/10.4161/cc.10.12.16011
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific postreplicative processing of mammalian telomeres. Science 2001; 293:2462-5; PMID:11577237; https://doi.org/10.1126/science.1062560
- d'Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol 2001; 11:1192-6; PMID:11516951; https://doi.org/10.1016/S0960-9822(01)00328-1
- Wang C, Lees-Miller SP. Detection and repair of ionizing radiation-induced DNA double strand breaks: new developments in nonhomologous end joining. Int J Radiat Oncol Biol Phys 2013; 86:440-9; PMID:23433795; https://doi.org/10.1016/j.ijrobp.2013.01.011
- Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem Cell Biol 2013; 91:31-41; PMID:23442139; https://doi.org/10.1139/bcb-2012-0058
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask MB, Streicher W, Jungmichel S, Nielsen ML, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 2015; 6:8088; PMID:26286827; https://doi.org/10.1038/ncomms9088
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 2015; 162:1066-77; PMID:26317470; https://doi.org/10.1016/j.cell.2015.07.047
- Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem 2013; 288:24731-41; PMID:23833192; https://doi.org/10.1074/jbc.M113.497974
- Krietsch J, Caron MC, Gagné JP, Ethier C, Vignard J, Vincent M, Rouleau M, Hendzel MJ, Poirier GG, Masson JY. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res 2012; 40:10287-301; PMID:22941645; https://doi.org/10.1093/nar/gks798
- Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, Frit P, Calsou P. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res 2014; 42:9047-62; PMID:25030905; https://doi.org/10.1093/nar/gku601
- Polo SE, Blackford AN, Chapman JR, Baskcomb L, Gravel S, Rusch A, Thomas A, Blundred R, Smith P, Kzhyshkowska J, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol Cell 2012; 45:505-16; PMID:22365830; https://doi.org/10.1016/j.molcel.2011.12.035
- Yuan M, Eberhart CG, Kai M. RNA binding protein RBM14 promotes radio-resistance in glioblastoma by regulating DNA repair and cell differentiation. Oncotarget 2014; 5(9):2820-6
- Yang D, Zhang ZJ, Oldenburg M, Ayala M, Zhang SC. Human embryonic stem cell-derived dopaminergic neurons reverse functional deficit in parkinsonian rats. Stem Cells 2008; 26:55-63, doi:2007-0494 [pii]10.1634/stemcells.2007-0494
- Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, O'Malley BW. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol Cell Biol 2004; 24:442-53; PMID:14673176; https://doi.org/10.1128/MCB.24.1.442-453.2004
- Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 2015; 210:529-39; PMID:26283796; https://doi.org/10.1083/jcb.201504117
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009; 33:717-26; PMID:19217333; https://doi.org/10.1016/j.molcel.2009.01.026
- Sasaki YT, Hirose T. How to build a paraspeckle. Genome Biol 2009; 10:227; PMID:19664169; https://doi.org/10.1186/gb-2009-10-7-227
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 2009; 106:2525-30; PMID:19188602; https://doi.org/10.1073/pnas.0807899106
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 2009; 19:347-59; PMID:19106332; https://doi.org/10.1101/gr.087775.108
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 2013; 16:1383-91; PMID:24036913; https://doi.org/10.1038/nn.3514
- Baechtold H, Kuroda M, Sok J, Ron D, Lopez BS, Akhmedov AT. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol Chem 1999; 274:34337-42; PMID:10567410; https://doi.org/10.1074/jbc.274.48.34337
- Bertrand P, Akhmedov AT, Delacote F, Durrbach A, Lopez BS. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene 1999; 18:4515-21; PMID:10442642; https://doi.org/10.1038/sj.onc.1203048
- Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gómez-Herreros F, Hafezparast M, Caldecott KW. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res 2014; 42:307-14; PMID:24049082; https://doi.org/10.1093/nar/gkt835
- Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol Biol 2012; 920:379-91; PMID:22941618
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 2009; 417:639-50; PMID:19133841; https://doi.org/10.1042/BJ20080413
- Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res 2013; 2:130-43; PMID:24000320
- Hammel M, Yu Y, Radhakrishnan SK, Chokshi C, Tsai MS, Matsumoto Y, Kuzdovich M, Remesh SG, Fang S, Tomkinson AE, et al. An intrinsically disordered APLF Links Ku, DNA-PKcs and XRCC4-DNA Ligase IV in an extended flexible non-homologous end joining complex. J Biol Chem 2016; 291(53):26987-7006
- Cottarel J, Frit P, Bombarde O, Salles B, Négrel A, Bernard S, Jeggo PA, Lieber MR, Modesti M, Calsou P. A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol 2013; 200:173-86; PMID:23337116; https://doi.org/10.1083/jcb.201203128
- Graham TG, Walter JC, Loparo JJ. Two-stage synapsis of DNA ends during non-homologous end joining. Mol Cell 2016; 61:850-8; PMID:26990988; https://doi.org/10.1016/j.molcel.2016.02.010
