ABSTRACT
The human mitochondrial chaperonin is a macromolecular machine that catalyzes the proper folding of mitochondrial proteins and is of vital importance to all cells. This chaperonin is composed of 2 distinct proteins, Hsp60 and Hsp10, that assemble into large oligomeric complexes that mediate the folding of non-native polypeptides in an ATP dependent manner. Here, we report the bacterial expression and purification of fully assembled human Hsp60 and Hsp10 recombinant proteins and that Hsp60 forms a stable tetradecameric double-ring conformation in the absence of co-chaperonin and nucleotide. Evidence of the stable double-ring conformation is illustrated by the 15 Å resolution electron microscopy reconstruction presented here. Furthermore, our biochemical analyses reveal that the presence of a non-native substrate initiates ATP-hydrolysis within the Hsp60/10 chaperonin to commence protein folding. Collectively, these data provide insight into the architecture of the intermediates used by the human mitochondrial chaperonin along its protein folding pathway and lay a foundation for subsequent high resolution structural investigations into the conformational changes of the mitochondrial chaperonin.
Introduction
The process of protein folding is critical for proper cellular function, yet remains one of the least understood biologic processes to date.Citation1,2 It is such an important function that nature has evolved a sophisticated macromolecular complex that specializes in folding proteins that cannot fold without assistance. Chaperonins are large oligomeric complexes that use the binding of ATP and the energy derived from its subsequent hydrolysis to trigger large conformational changes that guide the folding of nascent or misfolded proteins into their native state.Citation3 By providing kinetic assistance, chaperonins bind and sequester proteins inside a protected environment to allow proper protein folding and avoid irreversible protein aggregation and ultimately premature cell death.Citation4,5 It is estimated that 30% of cellular proteins require the aid of a chaperonin to assume their biologically active 3-dimensional structures.Citation6 The importance of chaperonins is best exemplified through knockout experiments revealing that the deletion of the host chaperonin gene is lethal in both bacteria and yeast.Citation7-10
Chaperonins are generally comprised of 14–18 subunits arranged as 2 stacked rings creating 2 internal chambers that can each sequester substrate proteins.Citation11 All chaperonins discovered thus far share a similar subunit architecture that consists of an apical domain capable of binding non-native proteins,Citation12 an equatorial domain that contains the ATP-binding pocket and major intra- and inter-ring contacts between subunits,Citation13 and a hinge-like intermediate region connecting the apical and equatorial domains.Citation14,15 Currently, chaperonins are categorized into 2 groups according to their oligomeric subunit organization and necessity for co-chaperonin to close-off the internal protein folding cavity.Citation16 However, the recent identification of a viral-encoded chaperonin that contains structural features from group I and group II provides sufficient evidence to warrant a distinct group of chaperonins.Citation17 In fact, there have been multiple studies over the past years that have identified single-ring chaperonins in a variety of organisms.Citation6,18-22 The difficulty in classifying single-ring chaperonins based on the current classification criteria warrants the introduction of a third group of chaperonins that is dependent on the utilization of single-ring intermediates.
The process of human mitochondrial protein folding is facilitated by a chaperonin that is currently classified as group I and is composed of the highly-conserved heat shock protein 60 and its co-chaperonin Hsp10.Citation23,24 Approximately 80% of Hsp60 is scattered throughout the mitochondria where it catalyzes the folding of proteins destined for the mitochondrial matrix.Citation3,25,26 The remaining Hsp60 can be found outside the mitochondria at locations such as the endoplasmic reticulum, the cell surface, peroxisomes and the cytosol.Citation27,28 The hsp60 gene HSPD1 is a nucleus-encoded gene that contains a mitochondrial targeting sequence that is necessary for successful import of the Hsp60 protein into mitochondria. The targeting sequence is then cleaved by mitochondrial proteases to yield a mature Hsp60 protein.Citation26 The Hsp60 protein that is not imported into the mitochondria retains the mitochondrial import signal and has been coined naïve Hsp60.Citation29
At present, it is unclear whether the mature or naïve Hsp60 protein constitutes the functionally active cytosolic Hsp60 due to conflicting conclusions from multiple studies. Immunoblotting experiments using an antibody against the mitochondrial targeting sequence of Hsp60 revealed that the naïve Hsp60 protein is located in the cytoplasm of rat liver cells and is imported into the mitochondria upon cellular stress.Citation30 Alternative experiments using mass spectrometry, N-terminal protein sequencing and immunochemistry have demonstrated that the mature Hsp60 protein is associated with the histone 2B (H2B) protein in the plasma membrane of CEM-SS T-cells.Citation31 It has also been postulated that both the mature and naïve Hsp60 protein may coexist in the mammalian cytoplasm.Citation29 In either case, a functionally compromised Hsp60/10 chaperonin complex in humans leads to mitochondrial dysfunction and impaired physiologic functions that have been deemed responsible for the onset of 2 distinct neurodegenerative diseases, as well as roles in early onset diabetes and juvenile rheumatoid arthritis.Citation32-34
Despite its biologic relevance, the labile nature of Hsp60 in-vitro has hindered structural and biochemical investigations. Consequently, functional details of Hsp60 and Hsp10 have largely been inferred from studies performed on the well-known bacterial homolog GroEL/ES. Although the amino acid identity between the mitochondrial chaperonin and its bacterial counterpart is high, the mitochondrial chaperonin operates via a single-ring intermediate indicating a novel protein folding mechanism distinct from the bacterial chaperonin system.Citation22,35 Thus, GroEL/ES by itself is not an ideal candidate for structural comparison to Hsp60 and a more appropriate chaperonin for comparison would be the newly reported phage-EL chaperonin.Citation17 The protein folding mechanism used by the Pseudomonas phage-EL chaperonin (phi-EL) is highlighted by its ability to dissociate from a double-ring complex into 2 single-rings. In addition, the inter-ring subunit organization and positive inter-ring cooperativity that are observed in phi-EL suggests that it utilizes a one stroke protein folding mechanism that relies on both rings being simultaneously active. In contrast, the GroEL/ES protein folding pathway is characterized by negative inter-ring cooperativity, where ATP binding in the cis-ring hinders ATP-binding in the trans-ring.Citation36 In spite of Hsp60s classification as a group I chaperonin, it is clear that Hsp60 shares structural features observed in both the phi-EL and GroEL/ES chaperonins. Therefore, a structural comparison against GroEL/ES and the phi-EL chaperonins would lead to a more accurate analysis of Hsp60.
Early studies on chimeric chaperonin proteins composed of the apical domain of GroEL and the equatorial domain from Hsp60 concluded that the resulting chimeric chaperonin folds denatured substrate solely as single-rings.Citation37 However, evidence for this conclusion did not include any structural data or data from wild-type Hsp60/10. Current literature has provided evidence that the human mitochondrial chaperonin likely uses both single and double-ring intermediates during its protein folding cycle.Citation38 Previous efforts have purified recombinant Hsp60 secreted from eukaryotic cells and as a fusion protein to glutathione S-transferase; while others have purified individual monomers that were then reassembled into oligomeric complexes upon the addition of ATP.Citation39-41 Such efforts were required due to the instability of Hsp60 in-vitro. Here, we present the bacterial expression and high-resolution purification of fully assembled and functional recombinant naïve Hsp60 and Hsp10 oligomeric complexes. Successful bacterial expression of fully assembled and functional Hsp60/10 chaperonin complex is likely due to the evolutionary relationship and similarity of mitochondria to bacteria. Although Hsp60 has been previously expressed in bacteria, our purification yields most the tetradecameric conformation of Hsp60 with very few monomers or single-ring conformations. Thus, our technique serves as a reliable method for the large-scale production of homogenous Hsp60 and Hsp10 proteins that can facilitate future biochemical and structural analyses.
Materials and methods
Hsp60 and Hsp10 protein expression and purification
The genes encoding the full-length wild type Hsp60 (HspD1) and Hsp10 (HspE1) proteins (with and without His6 tags) were cloned into the pET-30a expression vector system (EMD-Millipore, formally Novagen). The recombinant proteins were expressed in BL21 (DE3) E. coli cells (Invitrogen). Bacterial cells were cultured in 2xTY medium at 37°C for 4 hours until their density reached an absorbance between 0.6 and 0.8 at 600 nm. Protein expression was induced with IPTG (isopropyl-β-D-thiogalactopyranoside) at 30°C for an additional 4 hours. Cells were harvested by centrifugation at 5,000 xg for 30 minutes and lysed in 50 mM HEPES pH 7.5, 50 mM EDTA, 0.02% NaN3, in conjunction with treatment with hen egg white lysozyme (Sigma) and multiple freeze/thaw cycles. Lysates were treated with porcine liver DNase (Sigma) and 100 mM MgCl2. Saturated ammonium sulfate was then added to a final concentration of 50% (v/v), and the mixtures were incubated for one hour at 4°C. To purify the Hsp60 and Hsp10 proteins independently the ammonium sulfate precipitants were harvested by centrifugation and re-suspended in Buffer A (100 mM HEPES pH 7.5, 100 mM NaCl). A 500 μL aliquot of Hsp60 resuspension was diluted 10-fold against buffer A and loaded onto a Q-Sepharose 26/10 High Performance anion-exchange column that was pre-equilibrated with Buffer A and fractionated over a 0.1 M to 1 M NaCl linear gradient. Hsp60 eluted from the Q-Sepharose between 0.35 M and 0.45 M NaCl. The elutions of Hsp60 protein were then concentrated using a Vivaspin centrifugal concentrator with a 10 kDa molecular weight cut-off and loaded onto a Superose 6 size-exclusion column that was pre-equilibrated with Buffer A. Similarly, 500 µL of Hsp10 resuspension was diluted 10-fold in Buffer A and was loaded onto a Q-Sepharose anion exchange column. In contrast to Hsp60, Hsp10 was unable to bind to the Q-Sepharose and was found in the flow-through. The flow through was subsequently precipitated using 50% (m/v) ammonium sulfate and re-suspended in Buffer A. The re-suspended Hsp10 protein was then loaded on a Mono S 5/50 GL cation exchange column equilibrated with Buffer A and fractionated over a 0.1 M to 1 M NaCl linear gradient. Hsp10 eluted from the Mono-S column in the range of 0.5 M to 0.6 M NaCl. Sample homogeneity and the concentrations of both proteins were analyzed by 15% SDS-PAGE and bicinchoninic acid (BCA) protein assay, respectively. The approximate size of the oligomeric Hsp60 complex was identified on a 5% native polyacrylamide gel using β-galactosidase (Sigma) as a size control.
Dynamic light scattering
Dynamic light scattering (DLS) experiments were performed on the Malvern Zetasizer Nano-S instrument operated at 25°C. Purified APO Hsp60 and β-galactosidase (Sigma) were both diluted to a concentration of 0.025 mg/mL in Buffer A. A dust free disposable sizing cuvette was filled to 1.5 mL with the corresponding protein solution and measurements were taken every 3 seconds until 100 measurements were completed. The 100 measurements were averaged together to give a highly reproducible plot that more accurately identifies the hydrodynamic radius of the APO Hsp60.
ATPase activity
Before proceeding to structural work, the purified Hsp60 and Hsp10 proteins were tested for protein folding activity using the EnzChek Phosphatase Assay Kit (Molecular Probes, Leiden, The Netherlands) that measures inorganic phosphate released from enzymatic reactions through a spectrophotometric detection method. Reaction conditions recommended by the company were modified to accommodate the activity of our chaperonin. ATP and MgCl2 were added to a final concentration of 200 µM. The final concentration of Hsp60 in solution was 1 µM in a 200 µL reaction. Hsp10 was added at twice molar concentration of Hsp60 in both the positive and negative control reactions (2:1 molar ratio). The non-native protein substrate for Hsp60/10 was denatured α-lactalbumin that was denatured by the addition of EDTA to chelate bound Ca2+ and 50 mM DTT to reduce disulfide bonds. The mixture was heated to 98°C and allowed to cool before a 10-minute incubation period with the reaction mixture (200 µM MESG, 0.2 U Purine Nucleoside Phosphorylase (PNPase), Buffer A). A continuous colorimetric reading at 360 nm was initiated immediately after a mixture containing Hsp60 and Hsp10 were rapidly added to the reaction mixture.
Negative stain electron microscopy and single particle reconstruction
Negatively stained grids were prepared by applying 0.2 mg/mL of purified human mitochondrial APO-Hsp60 (with and without equimolar Hsp10) to continuous carbon film on 400-mesh copper grids that were glow discharged for 30 seconds. The excess protein solution was blotted using Whatman-1 filter paper and subsequent staining of the grids was done using 2% methylamine tungstate and 2% uranyl acetate.
The negative stain data set was collected on a JEOL 3200FS transmission electron microscope operated at 300 kV. CCD images were collected between 0.3–2.5 µm under-focus and at a magnification of 138,000X. The pixel size calculated for the data set was 1.09 Å/pixel. Processing of the micrographs was performed using the single particle analysis software “EMAN2”.Citation42 Particle selection was performed using the “e2boxer.py” program provided in the EMAN2 software package. A total of 2291 particles were picked from the micrographs. Following particle selection, CTF correction was performed using EMAN2s automated CTF fitting and the data was subsequently downsized by a factor of 2 using “e2ctf.py.” The downsized particles were then low pass and high pass filtered to 5 Å and 100 Å resolutions, respectively, using “e2ctf.py,” and then grouped according to their orientations and averaged together using the “e2refine2d.py” program to generate initial reference free class averages. The nicest class averages were then combined using the “e2initialmodel.py” program to reconstruct initial low-resolution 3-D models of the human mitochondrial Hsp60 in the APO conformation. The program “e2refinemulti.py” was used to separate the particles into 2 separate data sets. Finally, single map iterative refinements were performed using the 2-individual data sets using “e2refine_easy.py.” The iterative refinement process was continued until no further improvement in resolution occurred according to the Fourier Shell Correlation (FSC) curve. 1215 particles were used for the final reconstruction. The 3-D reconstructions were visualized using the UCSF-Chimera software package.Citation43 Docking of the Hsp60 crystal subunit (PDB 4PJ1) was performed using Chimera.Citation44
Results
Size determination of APO-Hsp60
To determine the relative size of the purified Hsp60 protein in its nucleotide and substrate free (APO) conformation we used dynamic light scattering (DLS), native gel electrophoresis, and negative stain electron microscopy. Aside from knowing that single and double-ring intermediates are part of the mitochondrial chaperonin protein folding pathway, the exact conditions that induce these oligomeric states are poorly understood. On the contrary, the APO conformations of the GroEL and the phage-EL chaperonins are characterized as double-ring, tetra-decameric complexes with approximate masses of 800 and 840 kDa, respectively.Citation15,45 Dynamic light scattering (DLS) measurements showed that our purified Hsp60 had a hydrodynamic diameter of 22 nm with a polydispersity index (PDI) of 0.209 (). β-galactosidase protein (Sigma) was used as a control for our DLS measurements since it also forms a large oligomeric complex that has an approximate mass of 464 kDa, and indicates whether APO-Hsp60 is in a tetradecameric or heptameric conformation. β-galactosidase protein displayed a smaller size of 18.8 nm and a PDI of 0.346 under our DLS experimental conditions.
Figure 1. Recombinant Hsp60 Protein Purity and Size Determination. a) 15% SDS-Gel depicting the purity and monomeric sizes of recombinant Hsp60 and Hsp10 proteins. Hsp10 runs anomalously due to the presence of a His6 Tag. b) 5% Native-Gel showing the purity and oligomeric size of the APO-Hsp60 protein and β-galactosidase protein (Sigma). c) Dynamic Light Scattering (DLS) of APO-Hsp60 (blue curve), 22.11 nm (PDI = 0.209); β-galactosidase (red curve), 18.76 nm (PDI = 0.346).
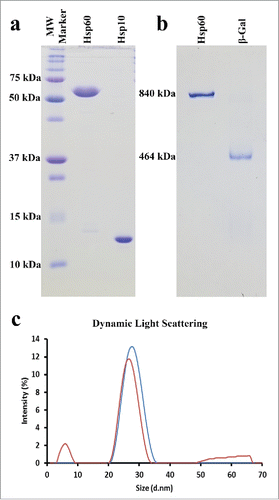
Native polyacrylamide gel electrophoresis can be used on non-denatured proteins and protein complexes to determine sample purity, heterogeneity, and the relative mass of native proteins and protein complexes. The results of our native gel show only one discernable protein band for Hsp60 with no detectable bands corresponding to contaminants, single-rings or other oligomeric Hsp60 complexes. The β-galactosidase protein is also well resolved as a single dominant band that was significantly lower in the gel than the APO-Hsp60 protein band seen in . The concentrations of the Hsp60 and β-galactosidase proteins in the native gel are approximately 7.5 µM and 4 µM, respectively.
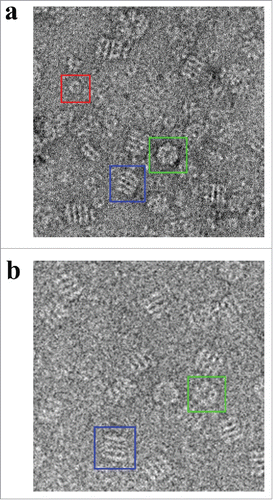
Under the electron microscope, Hsp60 displayed a conformation that is analogous to the APO conformation of GroEL ().Citation46,47 The observed Hsp60 particle orientations consisted of circular particles that displayed a 7-fold rotational subunit organization and rectangular particles that have 4 parallel striations as seen in the negative stain micrographs. Rectangular particles that consisted of 2 parallel striations were also observed; however, these particles were scarce and constituted less than 1% of the total particle count. Additionally, we were able to identify heptameric Hsp10 rings that were not associated with Hsp60.
Figure 2. Electron Micrographs of Hsp60 and GroEL. a) Negative stain electron micrograph showing APO-Hsp60 side views (blue box), Hsp60 top views (green box), and Hsp10 heptamers (red box). The particle contrast was inverted using EMAN2 so that the particles appear white on a dark background. b) Cryo-EM micrographs showing GroEL side views (blue box), and GroEL top views (green box). Figure was adapted and modified from Elad et al. (Citation2007).
Substrate initiates ATP hydrolysis in the human mitochondrial chaperonin
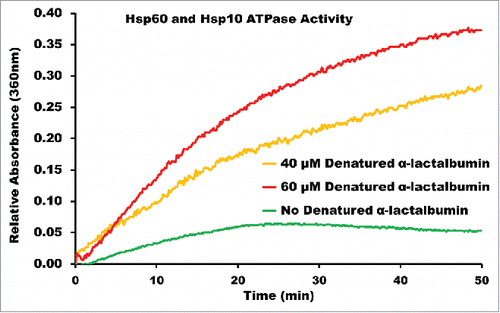
Purified Hsp60 and Hsp10 were incubated in the presence and absence of denatured α-lactalbumin supplemented with ATP to determine if the denatured substrate initiated ATP hydrolysis in the Hsp60/10 chaperonin as has been reported with other single-ring chaperonins.Citation17 The enzymatic hydrolysis of ATP by Hsp60 results in the release of inorganic phosphate that is then used by PNPase to convert 2-amino-6-mercapto-7methylpurine riboside (MESG) to ribose 1-phosphate and 2-amino-6-mercapto-7methylpurine, which has an absorbance that can be monitored at 360 nm. In the absence of substrate (), Hsp60/10 was unable to hydrolyze ATP and was dependent on the addition of non-native protein to stimulate ATPase activity.
Figure 3. ENZCHEK ATPase Assay of Recombinant Human Mitochondrial Hsp60 and Hsp10. Purified Hsp60 and Hsp10 in the presence of α-lactalbumin at 40 μM and 60 μM concentrations, and in the absence of α-lactalbumin. The rise in absorbance for Hsp60/10 without denatured α-lactalbumin can be attributed to self-hydrolysis of ATP over the time interval of the reaction.
Three-dimensional reconstruction of APO-Hsp60
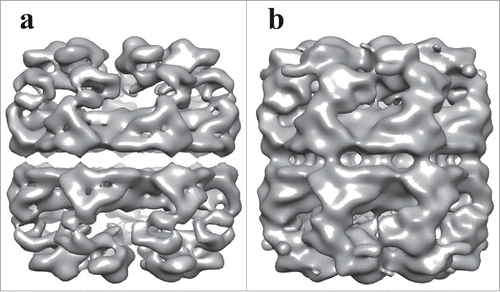
The individual APO-Hsp60 particles were picked from the negative stain micrographs using EMAN2 and were combined to generate a low resolution initial model of Hsp60 (), that was iteratively refined until convergence (). The observation of double-ring and presumably single-ring side views in the micrographs suggests heterogeneity in the sample, therefore EMAN2s version of multi-refine was used. After the data was split roughly in half, the double-ring particles were then used for a single map refinement that generated the structure seen in . The measured resolution from the FSC curve is approximately 14 Å at the 0.143 cut-off and 15 Å at the 0.5 cut-off, which is sufficient to resolve all 3 domains of the Hsp60 subunit. The dimensions of the reconstruction are 136 Å in diameter perpendicular to the 7-fold axis and 146 Å parallel to the long axis. A central slice perpendicular to the 7-fold axis reveals a planar equatorial domain connecting the 2 rings, while a central slice parallel to the 7-fold axis shows the protein folding cavities and the inter-ring space in the Hsp60 tetradecamer. The inter-ring subunits are arranged in a staggered (1:2) conformation that allows each Hsp60 subunit to contact 2 subunits in the opposite ring. The top-view of Hsp60 consisted of 7 radially arranged subunits that contacted each other at the apical domains. The features in the apical domains are well resolved at this resolution and feature a continuous collar of density connecting the apical domains. At a high iso-surface threshold tubular densities are observed, however, the connections between subunits appear broken (). While at a lower threshold, the tubular features are less resolved, but additional features such as an intra-ring subunit contact connecting the intermediate domain with the apical domain of the neighboring subunit can be better observed. An additional feature seen at a low iso-surface threshold is a contact connecting the equatorial and apical domain of the same subunit could be visualized (). The presented iso-surface threshold was chosen as the best display of all the structural elements resolved in the reconstruction.
Figure 4. Generation of the Initial Model of the Human Mitochondrial APO-Hsp60. a) Electron micrograph of APO-Hsp60 in the absence of Hsp10. Blue boxes encase Hsp60 side-views and green boxes encase top-views. b) APO-Hsp60 particles picked from the data set represented by the micrographs in “a.” c) Reference free initial class averages of the Hsp60 particles picked from the micrographs referenced in “b.” d) Hsp60 Initial model generated from the reference free class averages.
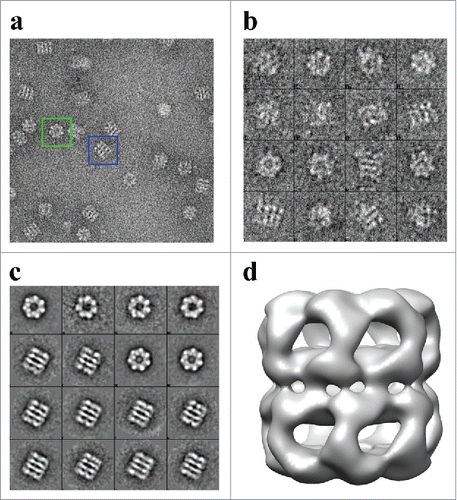
Figure 5. Iso-surface representation of the Human Mitochondrial APO-Hsp60 Reconstruction. a) Top view depicting the 7-fold arrangement of the Hsp60 subunits. b) The equatorial domains of the Hsp60 tetradecamer. c) Side-view orthogonal to the 7-fold axis showing the two heptameric rings that make-up the Hsp60 tetradecamer. d) Slab view displaying the interior of the tetradecamer and the inter-ring space. e) Fourier Shell Correlation (FSC) curve used to determine the resolution of the Hsp60 reconstruction at the 0.5 cutoff = 15 Å resolution, and the 0.143 cutoff = 14 Å resolution. The FSC curve shows that the data used for the reconstruction is self-consistent.
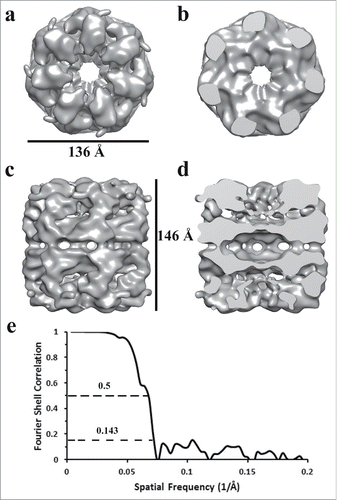
Figure 6. Comparison of Two Iso-Surface Thresholds of the APO-Hsp60 EM-Reconstruction. a) Side view of the Hsp60 reconstruction at a high iso-surface threshold. At high a threshold tubular densities can be observed but densities corresponding to intra- and inter-ring contacts appear broken. b) The Hsp60 reconstruction visualized at a lower iso-surface threshold. At a lower threshold, contacts between subunits are better resolved.
EMAN2 multi-refinement produced a second reconstruction from the presumed single-rings in the micrographs. The particles that consisted of 2 parallel striations were discarded during particle classification, possibly due to their low percentage, and the resulting reconstruction was generated from only circular particles. The lack of presumable single-ring side-views in the micrographs led to a reconstruction that did not resemble a chaperonin and therefore that sub data set was no longer used for any additional refinements.
Discussion
Initially, the Hsp60 and Hsp10 constructs were designed and expressed with N-terminal His6 tags to facilitate high yield purification via affinity column chromatography. Hsp60-His6 was found to bind to a HisTrap Ni-NTA column with notable affinity. However, subsequent size-exclusion chromatography revealed that the Hsp60-His6 protein did not assemble into an oligomeric quaternary structure required for functionality and was instead all monomers leading to the hypothesis that the His6 tag was likely preventing complex formation. Hsp60 was therefore subsequently cloned and expressed without the His6 affinity tag and was found to self-assemble within the bacterial cells into a functional tetradecameric structure. Hsp10 was also expressed with a His6 affinity tag but failed to bind to the HisTrap column. The His6 tag was likely inaccessible and did not bind to the Ni-NTA medium. The Hsp10 protein was expressed and purified through other chromatographic methods without the use of the His6 affinity tag. Despite Hsp60-His6 producing only monomers, Hsp10-His6 was able to assemble in to heptameric rings as observed in the negative stain electron micrographs indicating that the His6 tag had no effect on Hsp10 quaternary structure. It has been shown previously that purified Hsp60 and Hsp10 monomers require a pre-existing Hsp60/10 complex for assembly into their final quaternary structures.Citation48 Our results suggest that either no pre-existing chaperonin is required for assembling Hsp60 or that the GroEL/ES chaperonin from the BL21 E. coli cell line may be responsible in assisting the formation of the assembled Hsp60 and Hsp10 complexes that were purified.
An early investigation into the wild-type Hsp60 protein revealed its ability to function as a single toroidal ring.Citation22 The authors in this study used an ATP-agarose column to purify mature-Hsp60 with the bound protein being eluted upon the addition of 3 mM ATP. Therefore, it is plausible that the observed Hsp60 single-ring may have been a nucleotide induced conformation and is not reminiscent of APO-Hsp60. More recent structural investigations into the D3G and E321K mature-Hsp60 mutant proteins used His-tags to purify Hsp60 from bacterial cells.Citation44,49,50 However, following Hsp60 protein purification the His-tags were removed before in-vitro reconstitution of the oligomeric complexes for structural analysis. This was presumably done to avoid interference of the affinity tag during complex formation. An alternative structural investigation into the naïve-Hsp60 protein that was expressed in bacterial cells used His-tagged Hsp60 proteins that exhibited a dynamic equilibrium between tetradecameric and heptameric complexes at all concentrations assayed.Citation29 Our results indicate that the removal of the His6 tag from our recombinant, bacterially expressed Hsp60 protein allowed for the formation of a stable double-ring conformation with minimal traces of monomeric and single-ring Hsp60.
DLS on the purified Hsp60 protein expressed without the His6-tag confirmed the formation of the oligomeric complex. The resulting hydrodynamic diameter of 22 nm for Hsp60 was larger than the 18.8 nm diameter determined for β-Galactosidase (). The resulting differences in size indicated that our Hsp60 forms a substantially larger complex than the complex formed by β-galactosidase. The hydrodynamic diameter determined for our naïve-Hsp60 coincides well with previously published DLS results.Citation29
Further analysis of the size of APO-Hsp60 was performed using native gel electrophoresis. Like with DLS, we compared the size of the APO-Hsp60 oligomeric complex to the β-galactosidase complex. The physiologically active β-galactosidase protein forms a homo-tetrameric complex with an approximate mass of 465 kDa.Citation51 The corresponding protein band to the β-galactosidase complex appeared much lower on the gel than the protein band for Hsp60, suggesting that APO-Hsp60 was not in the single-ring conformation that is similar in mass to the β-galactosidase tetramer.
To determine if our purified Hsp60/10 chaperonin was biologically active, we monitored ATPase activity in the absence and presence of denatured α-lactalbumin. We found that the Hsp60/10 ATPase activity is completely dependent on the presence of denatured substrate, as seen in . The dependence on substrate for ATPase activity likely represents a physiologic “safety” mechanism built into the chaperonin preventing unwanted ATP hydrolysis while the chaperonin is not productively folding substrate protein. Recent studies in GroEL have proposed the symmetric GroEL/ES “football” intermediate to be the pre-dominant protein folding species and have also reported the dependence GroEL/ES's ATPase activity to initiate the conformational switch from the asymmetric “bullet” to the symmetric “football” shaped complexes.Citation52,53 Additionally, the recently identified chaperonin from the Pseudomonas phi-EL also displays substrate dependence for initiation of its protein folding mechanism.Citation17 Thus, it seems likely that substrate triggered ATP hydrolysis reflects an evolutionary conserved feature in all chaperonins. Furthermore, in the presence of large substrates such as denatured β-galactosidase (116 kDa), the 2 rings forming the tetradecameric conformation of the phi-EL chaperonin can dissociate into 2 heptameric single-rings that more than double the internal volume of the protein folding chambers. Both GroEL/ES and the phi-EL chaperonin require ATP binding and hydrolysis to induce allosteric changes that ultimately enable their protein folding abilities. The ATPase activity of Hsp60 indicates that this chaperonin is biologically active and is presumably transitioning through its various conformational intermediates during its folding of denatured α-lactalbumin. These results confirm that the human mitochondrial chaperonin is indeed dependent on the presence of non-native substrate to trigger ATPase activity and presumably protein folding activity. Further investigations are warranted to further understand how substrate-triggered ATP hydrolysis in the mitochondrial chaperonin affects the overall 3-D architecture.
Published purification methods have shown instability of Hsp60 tetradecameric complexes at low temperatures such as 4°C and dilute protein concentrations.Citation38,41,49 These previous Hsp60 purifications were performed at room temperature to avoid oligomeric dissociation. Our ability to purify APO-Hsp60 at 4°C and its subsequent imaging under the electron microscope in dilute concentrations further confirms the stability of our recombinant Hsp60 protein. The imaging of the homogeneous APO-Hsp60 complex using negative stain electron microscopy revealed the oligomeric assembly of Hsp60 to be in a double-ring tetradecameric conformation that was unable to bind its co-chaperonin Hsp10 in the absence of nucleotide (). This is confirmed in the reconstruction that clearly shows the apical domains of Hsp60 are in the closed conformation and incapable of binding substrate or Hsp10; features that are also observed in GroEL.Citation54 The 3-D reconstruction was generated using D7 symmetry indicating both rings are in the same conformational state. The approximate dimensions of the APO-Hsp60 reconstruction showed that it has a diameter of about 136 Å perpendicular to the 7-fold axis and about 146 Å parallel to the long axis of the particle, which coincide very well the dimensions of APO-GroEL and the recently published crystal structure of the E321K Hsp60 mutant.Citation15,44
The ability to generate a 3-D reconstruction demonstrates the purity and conformational stability of our APO-Hsp60 due to the fact that the single particle reconstruction method is highly dependent on the power of averaging.Citation55 Therefore, the features that are seen in the reconstruction are the resulting average of the constant features of all the particles included. The discernable features of negative stain reconstructions is usually limited to around 20 Å and is largely affected by the stain used.Citation56 Our combination of uranyl acetate and methylamine tungstate allowed for an intermediate resolution reconstruction with readily observable internal structural features. Fitting of the crystal subunit of the mature Hsp60 protein provides evidence that the particles used for the reconstruction were not distorted by the negative stain conditions. As seen in , an extra region of density is seen in the equatorial domain of our EM-reconstruction, where the C- and N-termini of Hsp60 can be located in the crystal subunit. The region of unfitted density in our map is near the termini of the crystal subunit and is likely the mitochondrial targeting sequence of naïve Hsp60. Another more obvious difference between the 2 structures can be seen in the apical domains (). The differences can be attributed to the different conformations of the 2 structures. In the crystal structure, the Hsp60/10 complex was formed by the addition of ATP, whereas our structure is of the co-chaperonin and nucleotide free Hsp60. In GroEL, the binding of nucleotide induces an elevation of the apical domains that allow the binding of co-chaperonin.Citation57 The structural differences observed between the APO-Hsp60 and ATP-Hsp60/10 structures are likely due to the nucleotide induced allosteric changes. The EM-reconstruction and ATPase assays provide strong evidence that the mitochondrial targeting sequence retained in the naïve Hsp60 protein does not affect the overall oligomeric structure or its protein folding activity.
Figure 7. Fitting of ATP-Hsp60/10 crystal structure into APO-Hsp60 EM-Reconstruction. a) Side view depicting the fitting between the crystal subunit (4PJ1) and the full Hsp60 EM-reconstruction. b) A single subunit showing the crystal subunit docked into the EM-Hsp60 subunit. The apical domain of the X-ray structure is in the ATP conformation and is therefore outside of the APO conformation density of the reconstruction. c) The subunit seen in “b” was rotated 180° and shows differences between the two maps. d) Side view of the APO-Hsp60 reconstruction showing the inter-ring subunit arrangement. The crystal subunit was adapted and modified from the 4PJ1 PDB structure.
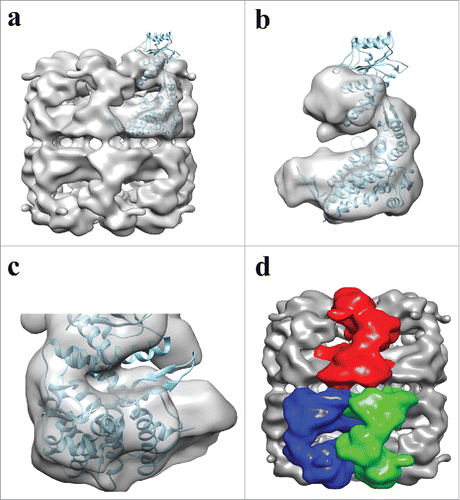
The mitochondrial chaperonin is reported to operate via a one-stroke mechanism that suggests a loss of negative co-operativity between the 2 rings.Citation22,37,58 In GroEL the subunits are arranged in an out-of-register (staggered) fashion with each monomeric subunit directly contacting 2 other subunits in the opposite ring.Citation15 This out-of-register inter-ring contact is directly responsible for the negative co-operativity between the 2 rings leading to the GroEL characteristic double stroke protein folding mechanism.Citation59 As in GroEL, APO-Hsp60 also displays an out-of-register inter-ring subunit organization with each Hsp60 subunit directly contacting 2 adjacent subunits in the opposite ring (). The out-of-register subunit organization between the 2 Hsp60 rings was first described in the mutant E321K crystal structure that showed each subunit directly contacts 2 subunits in the opposite ring. In addition, the contact surface area between the 2 rings in the E321K mutant is twice as extensive to that seen in GroEL, and was proposed that the enlarged contact surface may add stability to the oligomeric complexes of the mitochondrial chaperonin.Citation44 The extensive inter-ring contacts in the E321K structure could not be observed in our reconstruction. There are a few possibilities for this observation. First, the resolution of our reconstruction might not be adequate to distinguish all the inter-ring contacts in the Hsp60 tetradecamer. Second, our EM-reconstruction and the crystal structure are in completely different oligomeric conformations and the extensive contact interface may be a feature that is only present in the Hsp60/10/ATP complex. Third, since the Hsp60/10 chaperonin structure is dynamic the structural differences may be attributed to constraints on the proteins during crystallization, therefore, the enlarged contact surface may be an artifact induced during crystallization of the mutant Hsp60. High resolution crystal or cryo-EM structures are needed to shed light on the nature of the inter-ring contacts of the wild-type Hsp60/10 complexes. It is possible that the additional ring contacts reported for Hsp60 may allow for the transmittance of unique allosteric signals between the 2 Hsp60 rings leading to the one-stroke mechanism that has been reported for the mitochondrial chaperonin.Citation37,44,58
As mentioned previously, the mitochondrial chaperonin operates via a one-stroke protein folding mechanism that relies on the formation of single-ring intermediates. The single-ring phi-EL chaperonin displays an in-register inter-ring subunit organization and positive co-surgical inter-ring ATPase activity leading to a one-stroke protein folding mechanism.Citation17 The combination of these 2 features allows the phi-EL chaperonin to dissociate from a tetradecameric double-ring into 2 heptameric single-rings, similar to what is reported for the mitochondrial chaperonin. It is interesting to observe the out-of-register inter-ring contacts in Hsp60 despite its ability to function as a single-ring. Even though Hsp60 was initially reported to function as a single-ring, it is now clear that both single and double-ring intermediates are physiologic intermediates in the Hsp60/10 chaperonin protein folding cycle.Citation38,44 The unique structural features present in the phi-EL and the mitochondrial chaperonins that allow the use of double and single-ring intermediates reinforces the notion that a third group of chaperonins is necessary for classifying single-ring chaperonins. Future work is necessary to elucidate how the inter-ring contact arrangement used in the mitochondrial Hsp60/10 system dictates the formation of double and single-ring intermediates and if the inter-ring contacts are responsible for the proposed one-stroke mechanism seen in the mitochondrial chaperonin.Citation37
Collectively, these data indicate that the bacterial expression and purification of human mitochondrial naïve APO-Hsp60 yields an active and stable tetradecameric complex. The negative stain reconstruction presented here provides insight into the conformational stability of the Hsp60 tetradecamer and represents the first structure of the unaltered wild-type human mitochondrial Hsp60. Moreover, these results corroborate previous studies suggesting the use of double and single-ring intermediates in the protein folding pathway of various chaperonins, emphasizing the necessity for a third group of chaperonins. In conclusion, these data provide an impetus for further structural studies to elucidate the conditions that propel the human mitochondrial chaperonin through single- and double-ring intermediates along the protein folding cycle.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dr. Chuan Xiao and Jennie Choi for their assistance with the DLS experiments.
Funding
This work was made possible by the Welch Foundation award (AH-1649) and NIH-NIGMS (SC3GM113805) awarded to Ricardo A. Bernal. This work was supported by Grant 5G12MD007592 from the National Institutes on Minority Health and Health Disparities (NIMHD), a component of the National Institutes of Health (NIH).
References
- Ben-Naim A. Levinthal's question revisited, and answered. J Biomol Struct Dyn 2012; 30(1):113-24; PMID:22571437; https://doi.org/10.1080/07391102.2012.674286
- Bywater RP. Comments on the paper “Levinthal's question revisited, and answered” by A. Ben-Naim. J Biomol Struct Dyn 2013; 31(9):967-9; PMID:23297697; https://doi.org/10.1080/07391102.2012.748531
- Hildenbrand ZL, Bernal RA. Chaperonin-mediated folding of viral proteins. Adv Exp Med Biol 2012; 726, 307-24; PMID:22297519
- Ellis RJ. Molecular chaperones. Opening and closing the Anfinsen cage. Curr Biol 1994; 4(7):633-5.
- Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol 2007; 23, 115-45; PMID:17489689; https://doi.org/10.1146/annurev.cellbio.23.090506.123555
- Ferrer M, Lunsdorf H, Chernikova TN, Yakimov M, Timmis KN, Golyshin PN. Functional consequences of single:double ring transitions in chaperonins: life in the cold. Mol Microbiol 2004; 53(1):167-82; PMID:15225312; https://doi.org/10.1111/j.1365-2958.2004.04077.x
- Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol 1989; 171(3):1379-85; PMID:2563997; https://doi.org/10.1128/jb.171.3.1379-1385.1989
- Rospert S, Junne T, Glick BS, Schatz G. Cloning and disruption of the gene encoding yeast mitochondrial chaperonin 10, the homolog of E. coli groES. FEBS Lett 1993; 335(3):358-60; PMID:7903252; https://doi.org/10.1016/0014-5793(93)80419-U
- Reading DS, Hallberg RL, Myers AM. Characterization of the yeast HSP60 gene coding for a mitochondrial assembly factor. Nature 1989; 337(6208):655-9; PMID:2563898; https://doi.org/10.1038/337655a0
- Levy-Rimler G, Bell RE, Ben-Tal N, Azem A. Type I chaperonins: not all are created equal. FEBS Lett 2002; 529(1):1-5; PMID:12354603; https://doi.org/10.1016/S0014-5793(02)03178-2
- Zhang J, Baker ML, Schroder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, Frydman J, Chiu W. Mechanism of folding chamber closure in a group II chaperonin. Nature 2010; 463(7279):379-83; PMID:20090755; https://doi.org/10.1038/nature08701
- Fenton WA, Horwich AL. GroEL-mediated protein folding. Protein Sci 1997; 6(4):743-60; PMID:9098884; https://doi.org/10.1002/pro.5560060401
- Ranson NA, Farr GW, Roseman AM, Gowen B, Fenton WA, Horwich AL, Saibil HR. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell 2001; 107(7):869-79; PMID:11779463; https://doi.org/10.1016/S0092-8674(01)00617-1
- Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 1998; 93(1):125-38; PMID:9546398; https://doi.org/10.1016/S0092-8674(00)81152-6
- Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 1994; 371(6498):578-86; PMID:7935790; https://doi.org/10.1038/371578a0
- Reissmann S, Parnot C, Booth CR, Chiu W, Frydman J. Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat Struct Mol Biol 2007; 14(5):432-40; PMID:17460696; https://doi.org/10.1038/nsmb1236
- Molugu SK, Hildenbrand ZL, Morgan DG, Sherman MB, He L, Georgopoulos C, Sernova NV, Kurochkina LP, Mesyanzhinov VV, Miroshnikov KA, Bernal RA. Ring Separation Highlights the Protein-Folding Mechanism Used by the Phage EL-Encoded Chaperonin. Structure 2016; 24(4):537-46; PMID:26996960; https://doi.org/10.1016/j.str.2016.02.006
- Ishii N, Taguchi H, Sasabe H, Yoshida M. Equatorial split of holo-chaperonin from Thermus thermophilus by ATP and K+. FEBS Lett 1995; 362(2):121-5; PMID:7720857; https://doi.org/10.1016/0014-5793(95)00222-U
- Ishii N, Taguchi H, Sumi M, Yoshida M. Structure of holo-chaperonin studied with electron microscopy. Oligomeric cpn10 on top of two layers of cpn60 rings with two stripes each. FEBS Lett 1992; 299(2):169-74; PMID:1347504; https://doi.org/10.1016/0014-5793(92)80240-H
- Truscott KN, Hoj PB, Scopes RK. Purification and characterization of chaperonin 60 and chaperonin 10 from the anaerobic thermophile Thermoanaerobacter brockii. Eur J Biochem 1994; 222(2):277-84; PMID:7912671; https://doi.org/10.1111/j.1432-1033.1994.tb18866.x
- Semenyuk PI, Orlov VN, Sokolova OS, Kurochkina LP. New GroEL-like chaperonin of bacteriophage OBP Pseudomonas fluorescens suppresses thermal protein aggregation in an ATP-dependent manner. Biochem J 2016; 473(15):2383-93; PMID:27247423; https://doi.org/10.1042/BCJ20160367
- Viitanen PV, Lorimer GH, Seetharam R, Gupta RS, Oppenheim J, Thomas JO, Cowan NJ. Mammalian mitochondrial chaperonin 60 functions as a single toroidal ring. J Biol Chem 1992; 267(2):695-8. PMID:1346131
- Horwich A. Protein import into mitochondria and peroxisomes. Curr Opin Cell Biol 1990; 2(4):625-33; PMID:1979227; https://doi.org/10.1016/0955-0674(90)90103-L
- Dickson R, Larsen B, Viitanen PV, Tormey MB, Geske J, Strange R, Bemis LT. Cloning, expression, and purification of a functional nonacetylated mammalian mitochondrial chaperonin 10. J Biol Chem 1994; 269(43):26858-64; PMID:7929423
- Soltys BJ, Gupta RS. Immunoelectron microscopic localization of the 60-kDa heat shock chaperonin protein(Hsp60) in mammalian cells. Exp Cell Res 1996; 222(1):16-27; PMID:8549659; https://doi.org/10.1006/excr.1996.0003
- Singh B, Patel HV, Ridley RG, Freeman KB, Gupta RS. Mitochondrial import of the human chaperonin (HSP60) protein. Biochem Biophys Res Commun 1990; 169(2):391-6; PMID:1972619; https://doi.org/10.1016/0006-291X(90)90344-M
- Soltys BJ, Gupta RS. Cell surface localization of the 60 kDa heat shock chaperonin protein (hsp60) in mammalian cells. Cell Biol Int 1997; 21(5):315-20; PMID:9243807; https://doi.org/10.1006/cbir.1997.0144
- Soltys BJ, Gupta RS. Mitochondrial-matrix proteins at unexpected locations: are they exported? Trends Biochem Sci 1999; 24(5):174-7; PMID:10322429; https://doi.org/10.1016/S0968-0004(99)01390-0
- Vilasi S, Carrotta R, Mangione MR, Campanella C, Librizzi F, Randazzo L, Martorana V, Marino Gammazza A, Ortore MG, Vilasi A, et al. Human Hsp60 with its mitochondrial import signal occurs in solution as heptamers and tetradecamers remarkably stable over a wide range of concentrations. PLoS One 2014; 9(5):e97657; PMID:24830947; https://doi.org/10.1371/journal.pone.0097657
- Itoh H, Komatsuda A, Ohtani H, Wakui H, Imai H, Sawada K, Otaka M, Ogura M, Suzuki A, Hamada F. Mammalian HSP60 is quickly sorted into the mitochondria under conditions of dehydration. Eur J Biochem 2002; 269(23):5931-8; PMID:12444982; https://doi.org/10.1046/j.1432-1033.2002.03317.x
- Khan IU, Wallin R, Gupta RS, Kammer GM. Protein kinase A-catalyzed phosphorylation of heat shock protein 60 chaperone regulates its attachment to histone 2B in the T lymphocyte plasma membrane. Proc Natl Acad Sci U S A 1998; 95(18):10425-30; PMID:9724719; https://doi.org/10.1073/pnas.95.18.10425
- Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, Davoine CS, Brice A, Fontaine B, Gregersen N, Bross P. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet 2002; 70(5):1328-32; PMID:11898127; https://doi.org/10.1086/339935
- Magen D, Georgopoulos C, Bross P, Ang D, Segev Y, Goldsher D, Nemirovski A, Shahar E, Ravid S, Luder A, et al. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am J Hum Genet 2008; 83(1):30-42; PMID:18571143; https://doi.org/10.1016/j.ajhg.2008.05.016
- Henderson B, Fares MA, Lund PA. Chaperonin 60: a paradoxical, evolutionarily conserved protein family with multiple moonlighting functions. Biol Rev Camb Philos Soc 2013; 88(4):955-87; PMID:23551966; https://doi.org/10.1111/brv.12037
- Jindal S, Dudani AK, Singh B, Harley CB, Gupta RS. Primary structure of a human mitochondrial protein homologous to the bacterial and plant chaperonins and to the 65-kgdalton mycobacterial antigen. Mol Cell Biol 1989; 9(5):2279-83; PMID:2568584; https://doi.org/10.1128/MCB.9.5.2279
- Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 1997; 388(6644):741-50; PMID:9285585; https://doi.org/10.1038/41944
- Nielsen KL, Cowan NJ. A single ring is sufficient for productive chaperonin-mediated folding in vivo. Mol Cell 1998; 2(1):93-9; PMID:9702195; https://doi.org/10.1016/S1097-2765(00)80117-3
- Levy-Rimler G, Viitanen P, Weiss C, Sharkia R, Greenberg A, Niv A, Lustig A, Delarea Y, Azem A. The effect of nucleotides and mitochondrial chaperonin 10 on the structure and chaperone activity of mitochondrial chaperonin 60. Eur J Biochem 2001; 268(12):3465-72; PMID:11422376; https://doi.org/10.1046/j.1432-1327.2001.02243.x
- Zhou L, Xie J, Ruan Y, Zhu H, Wang W, Yun X, Guo L, Gan H, Sun L, Yu M, Gu J. Expression and purification of secreted recombinant hsp60 from eukaryotic cells. Protein Expr Purif 2010; 72(2):179-83; PMID:20362058; https://doi.org/10.1016/j.pep.2010.03.021
- Cunha DA, Zancope-Oliveira RM, Sueli M, Felipe S, Salem-Izacc SM, Deepe GS, Jr, Soares CM. Heterologous expression, purification, and immunological reactivity of a recombinant HSP60 from Paracoccidioides brasiliensis. Clin Diagn Lab Immunol 2002; 9(2):374-7; PMID:11874881
- Viitanen PV, Lorimer G, Bergmeier W, Weiss C, Kessel M, Goloubinoff P. Purification of mammalian mitochondrial chaperonin 60 through in vitro reconstitution of active oligomers. Methods Enzymol 1998; 290, 203-17; PMID:9534164
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 2007; 157(1):38-46; PMID:16859925; https://doi.org/10.1016/j.jsb.2006.05.009
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 2004; 25(13):1605-12; PMID:15264254; https://doi.org/10.1002/jcc.20084
- Nisemblat S, Yaniv O, Parnas A, Frolow F, Azem A. Crystal structure of the human mitochondrial chaperonin symmetrical football complex. Proc Natl Acad Sci U S A 2015; 112(19):6044-9; PMID:25918392; https://doi.org/10.1073/pnas.1411718112
- Roseman AM, Ranson NA, Gowen B, Fuller SD, Saibil HR. Structures of unliganded and ATP-bound states of the Escherichia coli chaperonin GroEL by cryoelectron microscopy. J Struct Biol 2001; 135(2):115-25; PMID:11580261; https://doi.org/10.1006/jsbi.2001.4374
- Roseman AM, Chen S, White H, Braig K, Saibil HR. The chaperonin ATPase cycle: mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell 1996; 87(2):241-51; PMID:8861908; https://doi.org/10.1016/S0092-8674(00)81342-2
- Elad N, Farr GW, Clare DK, Orlova EV, Horwich AL, Saibil HR. Topologies of a substrate protein bound to the chaperonin GroEL. Mol Cell 2007; 26(3):415-26; PMID:17499047; https://doi.org/10.1016/j.molcel.2007.04.004
- Cheng MY, Hartl FU, Horwich AL. The mitochondrial chaperonin hsp60 is required for its own assembly. Nature 1990; 348(6300):455-8; PMID:1978929; https://doi.org/10.1038/348455a0
- Parnas A, Nadler M, Nisemblat S, Horovitz A, Mandel H, Azem A. The MitCHAP-60 disease is due to entropic destabilization of the human mitochondrial Hsp60 oligomer. J Biol Chem 2009; 284(41):28198-203; PMID:19706612; https://doi.org/10.1074/jbc.M109.031997
- Nisemblat S, Parnas A, Yaniv O, Azem A, Frolow F. Crystallization and structure determination of a symmetrical ‘football’ complex of the mammalian mitochondrial Hsp60-Hsp10 chaperonins. Acta Crystallogr F Struct Biol Commun 2014; 70(Pt 1):116-9; PMID:24419632; https://doi.org/10.1107/S2053230X1303389X
- Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JL, Subramaniam S. 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science 2015; 348(6239):1147-51; PMID:25953817; https://doi.org/10.1126/science.aab1576
- Fei X, Ye X, LaRonde NA, Lorimer GH. Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form. Proc Natl Acad Sci U S A 2014; 111(35):12775-80; PMID:25136110; https://doi.org/10.1073/pnas.1412922111
- Ye X, Lorimer GH. Substrate protein switches GroE chaperonins from asymmetric to symmetric cycling by catalyzing nucleotide exchange. Proc Natl Acad Sci U S A 2013; 110(46):E4289-97; PMID:24167257; https://doi.org/10.1073/pnas.1317702110
- Clare DK, Vasishtan D, Stagg S, Quispe J, Farr GW, Topf M, Horwich AL, Saibil HR. ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin. Cell 2012; 149(1):113-23; PMID:22445172; https://doi.org/10.1016/j.cell.2012.02.047
- Frank J. Single-particle reconstruction of biological macromolecules in electron microscopy–30 years. Q Rev Biophys 2009; 42(3):139-58; PMID:20025794; https://doi.org/10.1017/S0033583509990059
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online 2004; 6, 23-34; PMID:15103397; https://doi.org/10.1251/bpo70
- Ranson NA, Clare DK, Farr GW, Houldershaw D, Horwich AL, Saibil HR. Allosteric signaling of ATP hydrolysis in GroEL-GroES complexes. Nat Struct Mol Biol 2006; 13(2):147-52; PMID:16429154; https://doi.org/10.1038/nsmb1046
- Nielsen KL, McLennan N, Masters M, Cowan NJ. A single-ring mitochondrial chaperonin(Hsp60-Hsp10) can substitute for GroEL-GroES in vivo. J Bacteriol 1999; 181(18):5871-5; PMID:10482535
- Sewell BT, Best RB, Chen S, Roseman AM, Farr GW, Horwich AL, Saibil HR. A mutant chaperonin with rearranged inter-ring electrostatic contacts and temperature-sensitive dissociation. Nat Struct Mol Biol 2004; 11(11):1128-33; PMID:15475965; https://doi.org/10.1038/nsmb844
