ABSTRACT
Accurate segregation of the duplicated genome in mitosis is essential for maintaining genetic stability. Errors in this process can cause numerical and/or structural chromosome abnormalities – hallmark genomic features commonly associated with both tumorigenesis and developmental disorders. A cell-based approach was recently developed permitting inducible missegregation of the human Y chromosome by selectively disrupting kinetochore assembly onto the Y centromere. Although this strategy initially requires several steps of genetic manipulation, it is easy to use, highly efficient and specific for the Y without affecting the autosomes or the X, and does not require cell cycle synchronization or mitotic perturbation. Here we describe currently available tools for studying chromosome segregation errors, aneuploidy, and micronuclei, as well as discuss how the Y-specific missegregation system has been used to elucidate how chromosomal micronucleation can trigger a class of extensive rearrangements termed chromothripsis. The combinatorial use of these different tools will allow unresolved aspects of cell division defects and chromosomal instability to be experimentally explored.
Introduction
Chromosome segregation errors during mitotic and meiotic cell divisions give rise to aneuploidy, an abnormal number of chromosomes. Aneuploidy can be frequently detected in the genome of cancer cellsCitation1 or individuals with developmental disorders, and is the leading cause of spontaneous miscarriages after fertilization. Emerging evidence also suggests a link between errors in mitosis and the formation of structural chromosomal translocations.Citation2 A primary cause of mitotic errors is the failure to correctly attach the spindle apparatus to kinetochores, large multi-protein complexes that assemble at the centromere of each chromosome and directly bind to spindle microtubules during mitosis.Citation3 Microtubule forces and microtubule-associated motors facilitate the movement of mitotic chromosomesCitation4 for alignment in metaphase and the physical separation of sister chromatids during anaphase, the stage of mitosis in which identical sets of chromosomes are segregated to each spindle pole to form the genomes of 2 daughter cells.
Errors during these processes can occur through several possible routes, some of which can escape surveillance by the mitotic checkpoint (or spindle assembly checkpoint), which safeguards against chromosome segregation defects: microtubules can fail to attach to one or both kinetochore(s) before anaphase onset, microtubules from the same pole can attach to both kinetochores of a sister chromatid pair (syntelic), or microtubules from both poles can attach to the same kinetochore of a sister chromatid pair (merotelic).Citation5 Each of these aberrant attachments can cause chromosomes to misalign in metaphase and/or lag behind during anaphase. Missegregated chromosomes can also become damaged during cytokinesisCitation2 or encapsulated into structures called micronuclei that are spatially isolated from the primary nucleus in interphase. Micronuclei were underappreciated for decades until recent sequencing efforts showed that complex structural rearrangements reminiscent of cancer-associated chromothripsisCitation6 could be formed on micronucleated chromosomes.Citation7,Citation8
Because chromosome segregation errors rarely occur in genetically stable human cells, several experimental tools are available to induce and/or investigate the complex effectsCitation9 of chromosome segregation errors, aneuploidy, and/or micronuclei. Here we review the advantages and limitations of current approaches, which can be broadly categorized into 2 primary types: those that provoke the missegregation of chromosomes at random versus those that are specific for either an artificial or authentic chromosome. Additionally, we describe the development of a recent method to selectively induce missegregation of the human Y chromosomeCitation10 – a strategy that has provided key insights into the mechanistic consequences of chromosome segregation errors and its role in driving chromothripsis.
Generating segregation errors at random
A spectrum of chemical inhibitors (summarized in ) can be used to trigger defects in mitosis and thereby increase the frequency of random chromosome segregation errors. One class of such inhibitors includes nocodazole and colcemid, which interferes with microtubule polymerization. These agents are typically added to an asynchronous cell population to first induce mitotic arrest through chronic activation of the mitotic checkpoint. Subsequent removal of the inhibitor allows re-polymerization of spindle microtubules but frequently causes one or a few kinetochores to form improper attachments. This procedure therefore causes one or a few chromosomes to missegregate in one or both daughter cells following the exit from mitosis. A non-chemical approach that similarly blocks microtubule polymerization is through transient exposure to cold temperatures.Citation11 On the opposite end, compounds such as taxol and its derivativesCitation12 stabilize microtubules, preventing the dynamic growth and shrinkage that are required for kinetochore attachment and correction of any attachment errors.Citation13 Other types of mitosis-arresting compounds include monastrolCitation14 and STLC (S-trityl-L-cysteine),Citation15 each of which inhibits the kinesin motor protein Eg5 and disrupts bipolar spindle formation,Citation16 or GSK923295,Citation17 which inhibits the CENP-E kinesin-like motor protein whose motive force is needed to power congression of initially misaligned chromosomes.Citation18
Table 1. Summary of available tools and approaches to study cell division defects and/or chromosome segregation errors.
Although these mitotic inhibitors are specific for their proposed targets, the efficiency of generating chromosome missegregation using these approaches is low due to the requirement for mitotic entry and arrest, requiring long inhibition incubation times as only ∼1–5% of cycling cell populations are actively in mitosis. Accumulated mitotic cells are then harvested (most easily by ‘mitotic shake-off’ or tapping of the culture dishes) and allowed to re-adhere in drug-free medium. A fraction of these cells subsequently develops one or more chromosome segregation errors as they exit from an extended mitosis and progress into G1. A large number of initial cells are therefore required to produce a small proportion of cells with missegregated chromosomes. Because cells are exposed to a prolonged mitosis, DNA damage and/or partial apoptotic responses can also be triggeredCitation19 even after completion of a normal, error-free division.Citation20
An alternative approach to arresting cells in mitosis is to instead override the mitotic checkpoint, which allows cells to prematurely proceed into anaphase before forming stable microtubule attachments to all kinetochores. Unlike agents that block mitosis, overriding the mitotic checkpoint is an efficient approach to produce segregation errors and does not require the use of cell cycle synchronization. Inhibitors of the Mps1 kinase, including reversineCitation21 or AZ3146,Citation22 are commonly used and can effectively cause premature anaphase onset in a broad number of cell types. The concentration of reversine can also be titrated to allow tight control over the severity of chromosome segregation errors such that low doses can trigger one error in a few cells and high doses can cause multiple errors in many cells.Citation23
These chemical approaches are commercially available, highly specific, easy to use, and applicable to a broad number of cell lines, thereby serving as valuable tools to study many aspects of normal and defective mitoses. A major disadvantage, however, is that these errors occur by chance and which chromosome undergoes missegregation (either into an incorrect daughter cell or a micronucleus) occurs at random. In the context of a diploid human cell, the probability of missegregating a specific autosomal chromosome-of-interest is ∼2 out of 46 (or ∼4.3%). Although random chromosome approaches can avoid potentially confounding chromosome-specific effects, it is particularly problematic for determining the fate of a missegregated chromosome since one cannot easily discriminate normally segregated vs. a missegregated chromosome in the immediate or subsequent cell cycles. Chromosomes sequestered in micronuclei are visually distinguishable during interphase until the nuclear envelope disassembles at the onset of mitosis. Approaches that can fluorescently label a specific chromosome or locus are possibleCitation24-28; however, significant cell-to-cell variability poses another challenge such that the chromosome in one micronucleus is different from another micronucleus in adjacent cells.
Avoiding randomness: Chromosome-specific approaches
The second category of approaches bypasses the specificity issue raised from chemical methods by targeting only a specific chromosome, including artificially created DNAs. Human artificial chromosomes (HACs) are ‘microchromosome-like’ DNA structures that span up to 10 megabases in length – one-fifth the size of the smallest human chromosome – that are created de novo or generated from a severely truncated chromosome. HACs have been engineered to harbor many features of authentic chromosomes, including a centromere, telomeres, and protein-coding genes, which can then be introduced, replicated, and maintained in host cells. Using the bacterial tetracycline operon/repressor system, HACs have also been designed with an inactivatable centromereCitation29,Citation30 that can be induced to missegregate, some of which subsequently enter micronuclei-like structures and acquire DNA damage.Citation31 HACs are excellent tools for studying chromosome missegregation rates,Citation32,Citation33 although they can become intrinsically unstable in the absence of selectionCitation34 and/or prone to unwanted recombination over an extended period. Whether the chromatin structure of HACs are similar to those of actual chromosomes has not been firmly established, but the sequence elements of artificially generated HACs do not generally reflect those typical of a human chromosome. Advances in synthetic chromosome technologies, as recently reported in yeast,Citation35 may also represent a possible next-generation class of artificial DNA tools.
Techniques involving microcell-mediated chromosome transfer (MMCT) have long been used to introduce HACs or entire (or parts of) genuine chromosomes from one cell into recipient host cells.Citation36 This is achieved by incorporating mitotic chromosomes into microcells (structures resembling micronuclei), in vitro purification and fusion of microcells with a host cell line through polyethylene glycol treatment, electrofusion, or with the use of Sendai viruses, and selection for a gene (i.e., a selectable marker or a fluorescent reporter) encoded by the transferred chromosome. MMCT permits studying the long-term effects of aneuploidy as it allows the generation of cells carrying additional chromosomes,Citation37-40 although the early consequences immediately following a chromosome missegregation event will almost certainly have been missed after the long experimental procedure to create the cells-of-interest. Like MMCT, Robertsonian translocations (caused by a fusion at or near the centromere between 2 acrocentric chromosomes to produce a single derivative chromosome carrying 2 non-homologous long arms) have also been cleverly exploited to generate trisomic mouse cell lines that harbor an additional copy of a defined chromosome.Citation41
A chromosome-specific missegregation strategy: Inactivating the Y centromere
An alternative approach involves missegregating a specific chromosome-of-interest through inactivation of its centromere – a specialized chromosomal locus designated for assembly of the kinetochore. Such an approach was originally established in Saccharomyces cerevisiae by forcing transcription through its nucleosome-sized point centromeres.Citation42,Citation43 By contrast, metazoans harbor megabase-long, heterochromatic centromeres that are epigenetically defined by the histone H3 variant Centromere Protein A (CENP-A). Centromere maintenance and function occurs through a 2-step mechanism.Citation44 In the first step, nascent CENP-A molecules are loaded into CENP-A-containing centromeric nucleosomes at the exit of mitosisCitation45 via its centromere-specific chaperone HJURP.Citation46,Citation47 In the second step, CENP-A utilizes its 2 terminal tails to directly and indirectly recruit CENP-C, the primary nucleator of the kinetochore,Citation44 to the centromere via functionally redundant mechanisms.
A gene replacement approach was recently developedCitation10 in an established human colorectal cancer cell line (DLD-1 cells with p53-inactivated) in which endogenous CENP-A proteins can be completely replaced by a chimeric CENP-A variant that does not support kinetochore assembly at centromeres lacking CENP-B (which binds to 17 base pair motif sequences called CENP-B boxes). Interestingly, only the Y chromosome centromere is deficient of CENP-B boxes; this is true in most of human genomes with rare exceptions being those harboring a neocentromere, as later discussed.
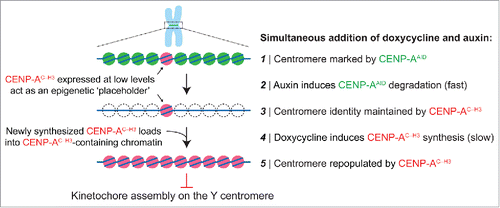
The replacement strategy requires at least 3 steps of genetic manipulation: 1) tagging the endogenous CENPA alleles with a ∼25 kDa auxin-inducible degron (AID) sequence [which can be achieved using standard genome editing technologies such as CRISPR/Cas9], 2) stable and high expression of the E3 ubiquitin ligase TIR1 [derived from the rice plant Oryza sativa], whose activation by indole-3-acetic acid (an auxin hormone) triggers ubiquitination and subsequent proteasome-mediated destruction of AID-tagged proteins, and 3) introduction of a doxycycline-inducible gene encoding the CENP-A chimera (called CENP-AC–H3) wherein the final 6 amino acids of CENP-A are swapped with the corresponding 3 amino acids from histone H3. This carboxy-tail chimera fails to support kinetochore formation at centromeres to which CENP-B is not bound.
Addition of doxycycline and auxin (both of which are non-toxic and cost effective) to the culture medium triggers CENP-A degradation and rescue with the chimeric variant, thereby producing an epigenetically marked yet non-functional Y centromere. Auxin-dependent degradation occurs much more rapidly (t1/2 of 9 minutes for removal of AID-tagged CENP-ACitation48) than centromeric chromatin replication, with wild-type or chimeric CENP-A loading through HJURP restricted to a 1–2 hour window after mitotic exit.Citation45 Pre-loading of the chimeric CENP-A at centromeres can be achieved by a low basal expression level due to intrinsic ‘leakiness’ of the promoter in the absence of doxycycline and/or by pre-induction with doxycycline for one or 2 cell cycle(s) before inducing auxin-mediated degradation of endogenous CENP-A (). Following doxycycline and auxin addition and CENP-A degradation, newly synthesized CENP-A chimeras are loaded into centromeric chromatin. As recent evidence has also indicated that CENP-A is not required for maintaining centromere function in mitosis after kinetochore assembly has been established during the prior interphase,Citation48 the rapid removal of CENP-A from centromeres leaves kinetochores intact on all centromeres except for the Y centromere (onto which a kinetochore had not assembled). As anticipated, this procedure efficiently triggers Y chromosome alignment failure and missegregation without extending the duration of mitosis.Citation10 Given that Y centromere position is maintained epigenetically by directed assembly of the CENP-A chimera, whether re-establishing endogenous CENP-A can cause centromere reactivation – and if so, to what extent – seems likely but has not been experimentally established.
Figure 1. Swapping out CENP-A at the centromere with an inducible gene replacement strategy. Gene editing is first used to add an auxin-inducible degron (AID) sequence to endogenous CENP-A, which undergoes rapid TIR1 E3 ubiquitin ligase-dependent degradation in the presence of the plant hormone auxin. Degraded CENP-AAID is then rescued by a doxycycline-inducible gene encoding a CENP-A/histone H3 carboxy-tail chimera (CENP-AC-H3) that does not support kinetochore assembly specifically on the Y centromere, thereby producing Y chromosome-selective segregation errors in the subsequent mitosis.
Although the strategy discussed here requires initial genetic manipulations, selective (and likely transient) centromere inactivation overcomes several limitations of currently available tools while retaining several key advantages. It neither requires nor causes arrest in mitosis as with microtubule-based drugs, and it is highly efficient in generating segregation errors in a large number of asynchronous cells as with Mps1 kinase inhibitors. This approach allows an authentic human chromosome to be studied as with MMCT, while also enabling the early events to be tracked over multiple cell cycles as with HAC-based systems. The Y is a bona fide chromosome spanning ∼57 megabases and represents the third smallest chromosome in the male human genome (it is larger than chromosomes 21 and 22) – over 5 times the size of the largest artificial HAC. Outside of XY pairing and recombination during male meiosis, the Y segregates through the same biological mechanisms as its autosomal counterparts in mitosis and is also transcriptionally active in somatic cultured cells (although only few genes are expressed).
Notwithstanding these advantages, there are also technical challenges involving the Y chromosome: only male cell lines can be used, and Y chromosome sequencing analysis can be complicated by regions of highly repetitive or palindromic sequences, as well as sequences sharing similarity to ones on the X chromosome. With the exception of a ∼30 megabase-long heterochromatic region on the distal Yq arm, however, the sequence of the euchromatic portion of Yp and Yq comprising ∼27 megabases has been determined to 99% completion.Citation49
The fate of missegregated chromosomes
Following centromere inactivation, missegregated Y chromosomes are subjected to several possible fates (). Because the Y is not essential for the growth of somatic cells in culture, whole-chromosome loss occurs most frequently, which may be avoided by introducing a selectable or fluorescent reporter into the Y. Chromosome gains also occur that produce daughter cells harboring multiple copies of the Y, although this is unlikely to trigger an aneuploidy-specific proteome imbalance responseCitation50 considering that the Y contains only ∼78 protein-coding genes.Citation49
Figure 2. A catastrophic fate for centromere-inactivated Y chromosomes. Following centromere inactivation using the CENP-A replacement strategy, the Y chromosome fails to properly congress during metaphase and subsequently missegregates in anaphase to produce aneuploid daughter cells with loss, gains, or micronucleation of the Y chromosome. When sequestered in a micronucleus, the Y chromosome undergoes extensive fragmentation in the next mitosis. The resulting fragments incorporate back into the nucleus for subsequent re-ligation by classical non-homologous end joining during interphase.
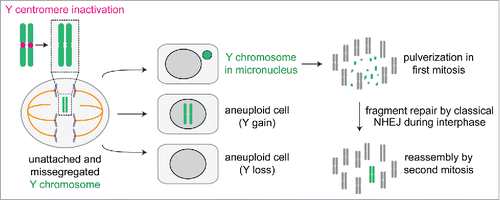
The most noteworthy fate for centromere-inactivated Y chromosomes is a significant enrichment for Y-containing micronuclei, the frequency of which peaks at ∼48 hours after addition of doxycycline and auxin. High-throughput sequencing of purified micronuclear DNAs revealed up to a 40-fold selective enrichment for the Y chromosome without changes to the autosomes or the X. Micronucleated Y chromosomes acquire DNA damage during interphase, consistent with micronucleation of random chromosomes,Citation31,Citation51 and subsequently undergo extensive fragmentation or shattering upon chromosome condensation in mitosis.Citation10,Citation52 These events give rise to multiple, distinct fragments that are subjected to DNA double-strand break repair in the subsequent interphase, the majority of which are re-ligated by classical non-homologous end joining. Use of this approach has therefore identified the stepwise mechanismsCitation10 contributing to how chromothripsis can be formed on initially micronucleated chromosomes.Citation7,Citation8
Future applications and closing remarks
Although the described centromere-specific inactivation approach is currently limited to the Y chromosome centromere that is inherently devoid of CENP-B boxes, the same strategy may also be applied to chromosomes containing an epigenetically active centromere at an otherwise non-centromeric region.Citation53 Over 100 examples of these ‘neocentromeres’ have been documented in the human context, and several cell lines have been established with a single chromosome harboring an active neocentromere with corresponding silencing of the original centromereCitation54 through an unknown mechanism. Because neocentromeres assembled onto 21 out of 22 autosomes and both sex chromosomes have been reported,Citation55,Citation56 it is in principle possible to create unique missegregation systems for all but one human chromosome. Like the Y centromere, these neocentromeres are deficient in CENP-B boxes and are therefore subjected to the same regulatory mechanisms for kinetochore assembly (that is, full dependence on the carboxy-terminal tail of CENP-A). One caveat is that many of the reported neocentromere-containing cell lines proliferate poorly in culture and may be difficult to genetically engineer. MMCT of the neocentromere-containing chromosome into an amenable host cell line may represent a feasible approach to develop chromosome-specific missegregation tools for autosomes.
Recognizing that there are multiple methods commonly used to generate chromosome segregation errors (with a particular focus here on chemical tools, artificial DNAs, and chromosome-specific centromere inactivation approaches), it is important to note that there is no ‘one size fits all’ method that is suitable for all experiments. Instead, the advantages of each approach should be exploited and likely used in combination to dissect the complex relationships between errors in cell division, aneuploidy, and structural chromosomal rearrangements. The cadre of tools available today, and those coming in the future, will permit detailed examination of both the short- and long-term consequences of chromosome segregation errors.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
D.W.C was supported by NIGMS (R01 GM074150 and R35 GM122476) and receives salary support from the Ludwig Institute for Cancer Research. P.L. was supported by a Hope Funds for Cancer Research Postdoctoral Fellowship (HFCR-14–06–06).
References
- Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumorigenesis. Cancer Cell 2008; 14:431-3; PMID:19061834; https://doi.org/10.1016/j.ccr.2008.11.011
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011; 333:1895-8; PMID:21960636; https://doi.org/10.1126/science.1210214
- Musacchio A, Desai A. A molecular view of kinetochore assembly and function. Biology (Basel) 2017; 6:pii: E5; PMID:28125021
- Walczak CE, Cai S, Khodjakov A. Mechanisms of chromosome behaviour during mitosis. Nat Rev Mol Cell Biol 2010; 11:91-102; PMID:20068571
- Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol 2008; 180:665-72; PMID:18283116; https://doi.org/10.1083/jcb.200712029
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011; 144:27-40; PMID:21215367; https://doi.org/10.1016/j.cell.2010.11.055
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature 2015; 522:179-84; PMID:26017310; https://doi.org/10.1038/nature14493
- Tan EH, Henry IM, Ravi M, Bradnam KR, Mandakova T, Marimuthu MP, Korf I, Lysak MA, Comai L, Chan SW. Catastrophic chromosomal restructuring during genome elimination in plants. Elife 2015; 4:pii: e06516
- Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 2015; 16:473-85; PMID:26204159; https://doi.org/10.1038/nrm4025
- Ly P, Teitz LS, Kim DH, Shoshani O, Skaletsky H, Fachinetti D, Page DC, Cleveland DW. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat Cell Biol 2017; 19:68-75; PMID:27918550; https://doi.org/10.1038/ncb3450
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol 2000; 10:59-67; PMID:10662665; https://doi.org/10.1016/S0960-9822(99)00276-6
- Schiff PB, Horwitz SB. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci U S A 1980; 77:1561-5; PMID:6103535; https://doi.org/10.1073/pnas.77.3.1561
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell 1986; 45:329-42; PMID:3516413; https://doi.org/10.1016/0092-8674(86)90318-1
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 1999; 286:971-4; PMID:10542155; https://doi.org/10.1126/science.286.5441.971
- DeBonis S, Skoufias DA, Lebeau L, Lopez R, Robin G, Margolis RL, Wade RH, Kozielski F. In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol Cancer Ther 2004; 3:1079-90; PMID:15367702
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 1992; 359:540-3; PMID:1406972; https://doi.org/10.1038/359540a0
- Qian X, McDonald A, Zhou HJ, Adams ND, Parrish CA, Duffy KJ, Fitch DM, Tedesco R, Ashcraft LW, Yao B, et al. Discovery of the First Potent and Selective Inhibitor of Centromere-Associated Protein E: GSK923295. ACS Med Chem Lett 2010; 1:30-4; PMID:24900171; https://doi.org/10.1021/ml900018m
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 1992; 359:536-9; PMID:1406971; https://doi.org/10.1038/359536a0
- Orth JD, Loewer A, Lahav G, Mitchison TJ. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol Biol Cell 2012; 23:567-76; PMID:22171325; https://doi.org/10.1091/mbc.E11-09-0781
- Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol 2010; 20:1666-71; PMID:20832310; https://doi.org/10.1016/j.cub.2010.08.018
- Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol 2010; 190:73-87; PMID:20624901; https://doi.org/10.1083/jcb.201001036
- Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, Green S, Taylor SS. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol 2010; 190:25-34; PMID:20624899; https://doi.org/10.1083/jcb.201002133
- Sansregret L, Patterson JO, Dewhurst S, Lopez-Garcia C, Koch A, McGranahan N, Chao WC, Barry DJ, Rowan A, Instrell R, et al. APC/C dysfunction limits excessive cancer chromosomal instability. Cancer Discov 2017; 7(2):218-33; PMID:28069571
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 1996; 135:1685-700; PMID:8991083; https://doi.org/10.1083/jcb.135.6.1685
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A 2015; 112:3002-7; PMID:25713381; https://doi.org/10.1073/pnas.1420024112
- Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, Pederson T. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 2016; 34:528-30; PMID:27088723; https://doi.org/10.1038/nbt.3526
- Anton T, Leonhardt H, Markaki Y. Visualization of genomic loci in living cells with a fluorescent CRISPR/Cas9 system. Methods Mol Biol 2016; 1411:407-17; PMID:27147056
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013; 155:1479-91; PMID:24360272; https://doi.org/10.1016/j.cell.2013.12.001
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 2008; 14:507-22; PMID:18410728; https://doi.org/10.1016/j.devcel.2008.02.001
- Kouprina N, Samoshkin A, Erliandri I, Nakano M, Lee HS, Fu H, Iida Y, Aladjem M, Oshimura M, Masumoto H, et al. Organization of synthetic alphoid DNA array in human artificial chromosome (HAC) with a conditional centromere. ACS Synth Biol 2012; 1:590-601; PMID:23411994; https://doi.org/10.1021/sb3000436
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012; 482:53-8; PMID:22258507; https://doi.org/10.1038/nature10802
- Lee HS, Lee NC, Grimes BR, Samoshkin A, Kononenko AV, Bansal R, Masumoto H, Earnshaw WC, Kouprina N, Larionov V. A new assay for measuring chromosome instability (CIN) and identification of drugs that elevate CIN in cancer cells. BMC Cancer 2013; 13:252; PMID:23694679; https://doi.org/10.1186/1471-2407-13-252
- Markossian S, Arnaoutov A, Saba NS, Larionov V, Dasso M. Quantitative assessment of chromosome instability induced through chemical disruption of mitotic progression. Cell Cycle 2016; 15:1706-14; PMID:27104376; https://doi.org/10.1080/15384101.2016.1175796
- Macnab S, Whitehouse A. Progress and prospects: human artificial chromosomes. Gene Ther 2009; 16:1180-8; PMID:19710706; https://doi.org/10.1038/gt.2009.102
- Zahn LM, Riddihough G. Building on nature's design. Science 2017; 355:1038-9; PMID:28280198
- Fournier RE, Ruddle FH. Microcell-mediated transfer of murine chromosomes into mouse, Chinese hamster, and human somatic cells. Proc Natl Acad Sci U S A 1977; 74:319-23; PMID:264685; https://doi.org/10.1073/pnas.74.1.319
- Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res 2004; 64:6941-9; PMID:15466185; https://doi.org/10.1158/0008-5472.CAN-04-0474
- Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, Storchova Z. The presence of extra chromosomes leads to genomic instability. Nat Commun 2016; 7:10754; PMID:26876972; https://doi.org/10.1038/ncomms10754
- Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesus M, Kantzler CL, Wangsa D, Barroso-Vilares M, Kale SD, Logarinho E, Cimini D. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci Rep 2016; 6:22828; PMID:26956415; https://doi.org/10.1038/srep22828
- Sheltzer JM, Ko JH, Replogle JM, Habibe Burgos NC, Chung ES, Meehl CM, Sayles NM, Passerini V, Storchova Z, Amon A. Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 2016; 31(2):240-55
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 2008; 322:703-9; PMID:18974345; https://doi.org/10.1126/science.1160058
- Chlebowicz-Sledziewska E, Sledziewski AZ. Construction of multicopy yeast plasmids with regulated centromere function. Gene 1985; 39:25-31; PMID:2934294; https://doi.org/10.1016/0378-1119(85)90103-9
- Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol 1987; 7:2397-405; PMID:3302676; https://doi.org/10.1128/MCB.7.7.2397
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol 2013; 15:1056-66; PMID:23873148; https://doi.org/10.1038/ncb2805
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 2007; 176:795-805; PMID:17339380; https://doi.org/10.1083/jcb.200701066
- Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 2009; 137:472-84; PMID:19410544; https://doi.org/10.1016/j.cell.2009.02.039
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 2009; 137:485-97; PMID:19410545; https://doi.org/10.1016/j.cell.2009.02.040
- Hoffmann S, Dumont M, Barra V, Ly P, Nechemia-Arbely Y, McMahon MA, Herve S, Cleveland DW, Fachinetti D. CENP-A is dispensable for mitotic centromere function after initial centromere/kinetochore assembly. Cell Rep 2016; 17:2394-404; PMID:27880912; https://doi.org/10.1016/j.celrep.2016.10.084
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003; 423:825-37; PMID:12815422; https://doi.org/10.1038/nature01722
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 2007; 317:916-24; PMID:17702937; https://doi.org/10.1126/science.1142210
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 2013; 154:47-60; PMID:23827674; https://doi.org/10.1016/j.cell.2013.06.007
- Kato H, Sandberg AA. Chromosome pulverization in human cells with micronuclei. J Natl Cancer Inst 1968; 40:165-79; PMID:5635016
- Scott KC, Sullivan BA. Neocentromeres: a place for everything and everything in its place. Trends Genet 2014; 30:66-74; PMID:24342629; https://doi.org/10.1016/j.tig.2013.11.003
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KH. Human centromere repositioning "in progress." Proc Natl Acad Sci U S A 2004; 101:6542-7; PMID:15084747; https://doi.org/10.1073/pnas.0308637101
- Burrack LS, Berman J. Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res 2012; 20:607-19; PMID:22723125; https://doi.org/10.1007/s10577-012-9296-x
- Amor DJ, Choo KH. Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet 2002; 71:695-714; PMID:12196915; https://doi.org/10.1086/342730
