ABSTRACT
Despite its ubiquity in interphase eukaryotic nuclei, the functional significance of the RabI configuration, in which interphase centromeres are clustered at the nuclear envelope (NE) near the centrosome and telomeres localize at the opposite end of the nucleus, has remained mysterious. In a broad variety of organisms, including Schizosaccharomyces pombe, the RabI configuration is maintained throughout mitotic interphase. The fission yeast linker of nucleoskeleton and cytoskeleton (LINC) complex mediates this centromere association. The functional significance of centromere positioning during interphase has been recently revealed using a conditionally inactivated LINC allele that maintains LINC stability but releases interphase centromere-LINC contacts. Remarkably, this interphase release abolishes mitotic spindle formation. Here, we confirm these observations using an alternative strategy to explore the role of centromere-NE association without modifying the LINC complex. We analyze spindle dynamics in cells lacking Csi1, a stabilizer of centromere-LINC associations, and Lem2, a NE protein harboring lamin interacting domains. We recapitulate these observations and their implications for the functional significance of centromere positioning for cell cycle progression in fission yeast and most likely, a wide range of eukaryotes.
Introduction
In the 1880s, studies performed by Carl Rabl in salamander larvae cells revealed that the chromosomal polarization imposed by anaphase, in which centromeres are pulled in opposite directions toward the poles and telomeres extend behind, persists in the following mitotic interphase.Citation1 This chromosome organization has since been known as the Rabl configuration.Citation2 The temporal persistence of the RabI configuration varies between species; in organisms such as S. pombe, it is maintained throughout interphase.Citation3 Despite the longevity of these observations, the function of the Rabl configuration has been a mystery. The absence of mutants able to fully disrupt the association of centromeres with the nuclear envelope (NE) beneath the spindle pole body (SPB, centrosome equivalent in yeast) during interphase while preserving intact kinetochores and SPBs has thwarted our ability to assess the relevance of centromere positioning.
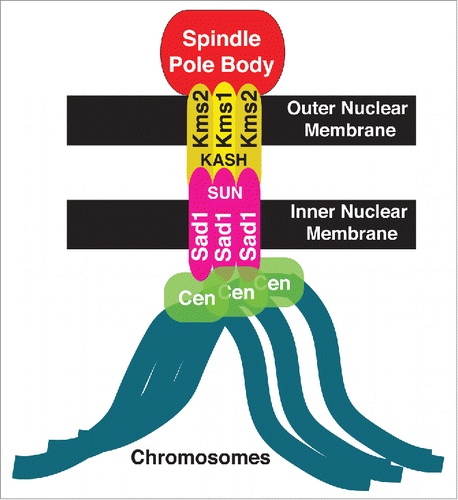
These problems were solved recently. Centromere-NE associations are mediated by the LINC complex, which consists of the SUN-domain protein Sad1, lodged in the inner nuclear membrane, and its outer nuclear membrane spanning KASH domain partners Kms1 and Kms2Citation3,4 (). The development of a new thermo-sensitive allele of Sad1, Sad1.2, in which centromere-LINC contacts are inactivated at 36°C while Sad1 nonetheless remains stable at the SPB, has disclosed a crucial role of centromere-LINC associations as a controller of spindle assembly.Citation5 This control is exerted via regulation of the partial NE breakdown required for SPB insertion into the NE, a crucial prerequisite for spindle assembly.
Figure 1. Schematic of the LINC complex in fission yeast. The LINC complex consists of 2 components: Sad1, the SUN-domain protein (pink) in the inner nuclear membrane and a KASH-domain protein (yellow) in the outer nuclear membrane. In fission yeast, Sad1 anchors all centromeres to the inner nuclear membrane.
Although S. pombe is considered to undergo a closed mitosis in which the spindle forms within an intact NE, the SPB sits on the cytoplasmic face of the NE throughout interphase. Hence, to nucleate a spindle in the compartment harboring the chromosomes, the NE must be locally disassembled beneath the SPB at mitotic onset. This crucial step generates a NE hole known as a fenestration. Once the SPB is duplicated, the 2 SPBs descend into this fenestration and form the spindle. This localized and transient NE disassembly in fission yeast is reminiscent of the nuclear envelope breakdown (NEBD) process in metazoans, in which the entire NE is disassembled to license the access of the spindle to the chromosomes.Citation6 Release of all centromeres from their normal LINC association during interphase, via inactivation of the LINC complex using the Sad1.2 allele, abolishes NE fenestration.Citation5 The conservation of NE disassembly, albeit to differing extents, as well as the LINC complex and the centromere components, raises the possibility that centromere-LINC contacts are a conserved controller of NE disassembly.
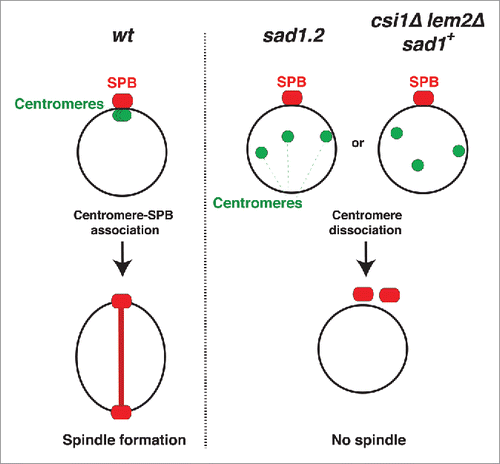
The completeness of the linkage between centromeres and Sad1 is mediated by Csi1 (chromosome segregation impaired 1), a Sad1-interacting protein identified in a screen for factors required for proper chromosome segregation.Citation7 Deletion of csi1 confers centromere clustering defects in ∼80% of cells; however, in all csi1Δ cells at all times, at least one centromere remains associated with Sad1.Citation7 The penetrance of csi1Δ defects is increased upon loss of the inner nuclear membrane protein Lem2, suggesting that Csi1 and Lem2 cooperate in linking centromeres to the LINC complex during interphase. Indeed, a small subset of still images of csi1Δ lem2Δ cells show all centromeres dissociated from the SPB.Citation8
We found that the interaction of only one centromere with the LINC complex during interphase is sufficient to confer proper NE disassembly and spindle formation in mitosis and meiosis.Citation5,Citation9 These observations provide a potential explanation for the overall lack of spindle defects in csi1Δ lem2Δ cells. If the total centromere-LINC dissociation seen in some csi1Δ lem2Δ cells were transient, the residual centromere-LINC contact level would be permissive for spindle formation. Conversely, if simultaneous loss of Csi1 and Lem2 were to occasionally confer irreversible and complete centromere dissociation, our work predicts events of spindle failure. To examine these possibilities, we examined the behavior of csi1Δ lem2Δ cells using live microscopy. We find that while centromere dissociation induced by sad1.2 mutations is irreversible, the association between centromeres and Sad1 in a csi1Δ lem2Δ background is destabilized rather than severed; dissociated centromeres often return to the SPB this scenario. Nevertheless, we observe a low frequency of csi1Δ lem2Δ cells with all centromeres persistently dissociated from the LINC complex. In this subset of cells, similar spindle formation defects to those observed in sad1.2 cells at restrictive temperatures are observed. These observations confirm that centromere localization per se, whether in a sad1+ or sad1-deficient background, is required for spindle nucleation.
Results
Transient centromere-LINC associations in csi1Δ lem2Δ cells
We used time-lapse imaging to quantify the number of cells in which all centromere foci are separated from the SPB () during the 40 min before mitotic SPB duplication, when cells are in interphase. To visualize the position of the centromeres with respect to the interphase SPB, we endogenously tagged a centromeric protein, Mis6, with GFP and a SPB component, Sid4, with mCherry; tubulin was visualized via ectopic expression of mCherry-Atb2 to identify cells in interphase. In wt cells, all centromere signals localize to a single focus beneath the interphase SPB (−30 min to −10 min in ). In csi1Δ cells, this centromere clustering is disrupted; although at least one centromere localizes to the SPB, additional Mis6-GFP foci are seen elsewhere (7; −50 min to −10 min in ). Deletion of lem2 in csi1Δ cells confers greater defects in centromere clusteringCitation8; some csi1Δ lem2Δ cells show all centromere foci separated from the interphase SPB (−60 min in ). In most instances, dissociated Mis6 foci return to the SPB region during interphase (−40 min to −10 min in ). This transient centromere-SPB association differs from the irreversible centromere dissociation induced by sad1.2 cells at restrictive temperature (Citationref. 5; ). To analyze instances of total centromere dissociation (ie, excluding cells with one or more centromere(s) remaining beneath the SPB throughout interphase), we allocated cells into 2 categories: (1) transient total dissociation, in which centromeres return to the LINC complex region beneath the SPB during the 40 minutes preceding SPB duplication, and (2) persistent total dissociation, in which centromeres fail to return to the SPB. After 4 hours at restrictive temperature, ∼18% of sad1.2 cells show persistent total dissociation (5; ) while transient total dissociation is never seen (). In contrast, ∼10% of csi1Δ lem2Δ cells show transient total dissociation while only ∼1% maintain persistent total dissociation ( and ). Therefore, Csi1 and Lem2 redundantly stabilize centromere associations with Sad1, whose function is absolutely required to anchor centromeres to the region.
Figure 2. Persistent total centromere dissociation in csi1Δ lem2Δ cells results in spindle failure. (A) In wt cells, all centromeres are clustered beneath the SPB during mitotic interphase. Double deletion of csi1 and lem2, or sad1.2 cells grown at restrictive temperature (36°C), lead to centromere dissociation from the SPB during interphase. (B) Series of frames of films of mitotic cells harboring Sid4-mCherry (SPB), Mis6-GFP (centromeres) and ectopically expressed mCherry-Atb2 (Tubulin). Bars, 5 µm. Numbers indicate mitotic progression in minutes; t = 0 is the SPB duplication stage. Yellow arrowheads point to SPBs failing to separate. (C-D) Quantitation of centromere dissociation exemplified in (B). Total centromere dissociation of the centromeres from the SPB is never observed in wt or csi1Δ cells. sad1.2 grown for 4h at 36 °C commence total centromere dissociation from the SPB.
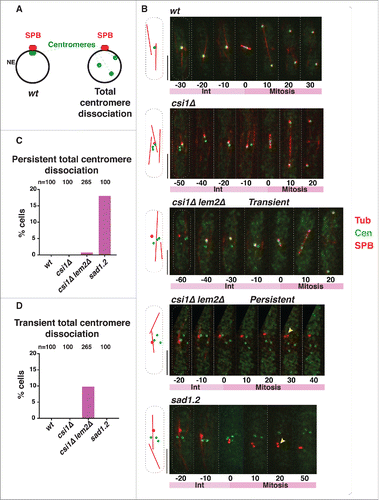
Persistent centromere dissociation in csi1Δ lem2Δ cells abolishes spindle formation
The occasional occurrence of persistent total centromere-LINC dissociation by simultaneous deletion of csi1 and lem2 provides the opportunity to determine whether loss of centromere contacts abolishes spindle formation even if Sad1 is fully intact (sad1+). In wt cells, the SPB duplicates before mitosis, during which the elongating spindle separates the 2 SPBs (). The partial centromere dissociation observed in csi1Δ cells does not affect spindle formation (); this is in accord with our observations that a single centromere-SPB contact confers proper spindle assembly.Citation5,Citation9 Those csi1Δ lem2Δ cells showing transient total dissociation are also able to promote spindle formation (−40 min to −10 min in ). However, those csi1Δ lem2Δ cells showing persistent total dissociation accomplish SPB duplication but fail to separate the duplicated SPBs (30 min in , yellow arrow); this behavior mirrors that of sad1.2 cells at restrictive temperature (20 min , yellow arrow). Hence, the complete loss of centromere-Sad1 contacts, even in the context of a wt Sad1 protein, confers spindle failure, thus confirming that the spindle failure seen in sad1.2 cells stems specifically from centromere release ().
Discussion
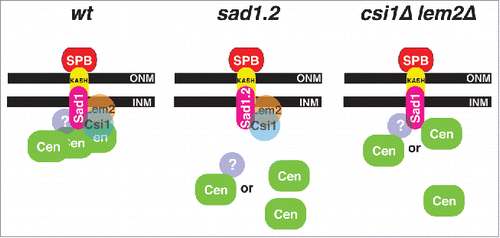
The Sad1-interacting proteins Csi1 and Lem2 reinforce centromere positioning beneath the SPB during interphase.Citation7,Citation8,Citation10,Citation11 While the loss of Csi1 leads to penetrant defects in the persistence of centromere clustering,Citation7 centromere-LINC associations are largely intact in lem2Δ cells.Citation8,Citation10 Models for how centromeres are connected with the LINC complex need to account for the facts that most csi1Δ lem2Δ cells show at least one centromere-LINC association, and that Csi1 and Lem2 remain stably associated with Sad1.2 at restrictive temperatures despite the irreversible loss of centromere associations.Citation5 At least two, not mutually exclusive, possibilities can accommodate our observations thus far. First, a still unidentified third factor may mediate centromere-LINC associations in a Sad1-dependent but largely Csi1/Lem2 independent manner. Second, kinetochore proteins might interact with Sad1 directly, with Csi1 and Lem2 stabilizing this interaction to different degrees (). Such stabilization could be via interactions between Csi1/Lem2 and the kinetochores, via conformational alteration of Sad1 by Csi1/Lem2, or via conformational alteration of kinetochores by Csi1/Lem2. Sad1.2 harbors two substitutions: threonine to serine at position 3 and serine to proline at position 52,Citation5 both of which are required for the sad1.2 phenotype. These mutations may lead to a conformational change in the Sad1 N-terminal domain that compromises interaction with the ‘third’ element, be it a kinetochore factor or an additional connecting factor. Current genome-wide screens aim to identify those factors involved in connecting centromeres to the LINC complex.
Figure 4. Schematic of centromere-LINC organization during interphase. Proposed organization in wt, sad1.2, and csi1Δ lem2Δ cells; see text for details. INM, inner nuclear membrane; ONM, outer nuclear membrane.
Spindle failure in the absence of centromere-LINC contacts is preceded by failure of NE disassemblyCitation5; indeed, this failure alone can account for the role of centromere-LINC contacts in promoting spindle formation.Citation6 How might centromeres impinge on NE fenestration? Although the mechanisms governing NE disassembly remain unclear, it has been observed that disassembly of NE-associated components, such as the nuclear pore complexes or nuclear lamina, is integral to NEBD and requires precise phosphorylation events. These modifications are coordinately catalyzed by the activity of cyclin-dependent kinase (CDK1), polo-like kinase (PLK1) and Aurora B. The ability of centromeres to recruit enzymes like CDK1 and PLK1Citation6,12 suggests the possibility that centromere-LINC interactions might concentrate these enzymes at the NE to trigger its disassembly. Alternatively or in addition, centromeres might recruit membrane wedging proteins to the region. We are currently exploring these ideas to discern how specific regions of the chromosome interface with the NE remodeling and cell cycle control machineries. These control modules may provide coupling between events within and outside of the nucleus, ensuring that NEBD and spindle assembly cannot occur prematurely.
Methods
Strains and growth conditions
Strains used in this work are listed in . Standard fission approachesCitation13 were performed. Gene C-terminal tagging was performed as described.Citation9,Citation14 Insertions of mCherry-Atb2 at the aur1 locusCitation15 used pYC19-mCherryAtb216 provided by T. Toda (Hiroshima University). Final concentrations of aureobasidin A (0.5 µg/mL), nourseothricin (100 µg/mL clonNAT), G418 (150 µg/mL geneticin), hygromycin B (300 µg/mL), thiabendazole (10–15 µg/mL TBZ) were used for selection.
Table 1. Strains constructed and analyzed in this work.
Fluorescence microscopy and live analysis
Fluorescence microscopy data were obtained using the DeltaVision microscope system (Applied Precision, Seattle, WA). Cells were adhered to 35 mm glass culture dishes (MatTek) using 0.2 mg/ml soybean lectin (Sigma) and immersed in EMM (with required supplements). Time-lapse imaging was performed at 30°C and 36°C in an Environmental Chamber with a DeltaVision Spectris (Applied Precision) comprising an Olympus IX70 wide field inverted epifluorescence microscope, an Olympus UPlanSapo 100x NA 1.4 oil immersion objective, and a Photometrics CCD CoolSnap HQ camera. Images were acquired over 26 focal planes at a 0.35 μm step size. Images were deconvolved and combined into a 2D image using the maximum intensity projection setting using SoftWorx (Applied Precision).
Abbreviations
| LINC | = | linker of nucleoskeleton and cytoskeleton |
| NE | = | nuclear envelope |
| NEBD | = | nuclear envelope breakdown |
| SPB | = | spindle pole body |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank our lab members for discussions, and Ramon R. Barrales and Sigurd Braun for csi1 lem2 strains.
Funding
This work was supported by the National Institutes of Health.
References
- Rabl C. Uber Zellteilung. Morphologisches Jahrbuch 1885; 10:214-330.
- Cowan CR, Carlton PM, Cande WZ. The polar arrangement of telomeres in interphase and meiosis. Rabl organization and the bouquet. Plant Physiol 2001; 125:532-8; PMID:11161011; https://doi.org/10.1104/pp.125.2.532
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol 1993; 121:961-76; PMID:8388878; https://doi.org/10.1083/jcb.121.5.961
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Developmental Cell 2009; 17:598-605; PMID:19922865; https://doi.org/10.1016/j.devcel.2009.10.014
- Fernandez-Alvarez A, Bez C, O'Toole ET, Morphew M, Cooper JP. Mitotic nuclear envelope breakdown and spindle nucleation are controlled by interphase contacts between centromeres and the nuclear envelope. Dev Cell 2016; 39:544-59; PMID:27889481; https://doi.org/10.1016/j.devcel.2016.10.021
- Fernandez-Alvarez A, Cooper JP. Chromosomes orchestrate their own liberation: Nuclear envelope disassembly. Trends Cell Biology 2017; 27:255-265.
- Hou H, Zhou Z, Wang Y, Wang J, Kallgren SP, Kurchuk T, Miller EA, Chang F, Jia S. Csi1 links centromeres to the nuclear envelope for centromere clustering. J Cell Biol 2012; 199:735-44; PMID:23166349; https://doi.org/10.1083/jcb.201208001
- Barrales RR, Forn M, Georgescu PR, Sarkadi Z, Braun S. Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev 2016; 30:133-48
- Fennell A, Fernandez-Alvarez A, Tomita K, Cooper JP. Telomeres and centromeres have interchangeable roles in promoting meiotic spindle formation. J Cell Biol 2015; 208:415-28; PMID:25688135; https://doi.org/10.1083/jcb.201409058
- Hiraoka Y, Maekawa H, Asakawa H, Chikashige Y, Kojidani T, Osakada H, Matsuda A, Haraguchi T. Inner nuclear membrane protein Ima1 is dispensable for intranuclear positioning of centromeres. Genes Cells 2011; 16:1000-11; PMID:21880100; https://doi.org/10.1111/j.1365-2443.2011.01544.x
- Tange Y, Chikashige Y, Takahata S, Kawakami K, Higashi M, Mori C, Kojidani T, Hirano Y, Asakawa H, Murakami Y, et al. Inner nuclear membrane protein Lem2 augments heterochromatin formation in response to nutritional conditions. Genes Cells 2016; 21:812-32; PMID:27334362; https://doi.org/10.1111/gtc.12385
- Decottignies A, Zarzov P, Nurse P. In vivo localisation of fission yeast cyclin-dependent kinase cdc2p and cyclin B cdc13p during mitosis and meiosis. J Cell Sci 2001; 114:2627-40; PMID:11683390
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 1991; 194:795-823; PMID:2005825
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998; 14:943-51; PMID:9717240; https://doi.org/10.1002/(SICI)1097-0061(199807)14:10%3c943::AID-YEA292%3e3.0.CO;2-Y
- Hashida-Okado T, Yasumoto R, Endo M, Takesako K, Kato I. Isolation and characterization of the aureobasidin A-resistant gene, aur1R, on Schizosaccharomyces pombe: roles of Aur1p+ in cell morphogenesis. Curr Genetics 1998; 33:38-45; PMID:9472078; https://doi.org/10.1007/s002940050306
- Nakamura Y, Arai A, Takebe Y, Masuda M. A chemical compound for controlled expression of nmt1-driven gene in the fission yeast Schizosaccharomyces pombe. Analytical Biochem 2011; 412:159-64; PMID:21295003; https://doi.org/10.1016/j.ab.2011.01.039