ABSTRACT
Increasing attention has been paid to certain ribosomal or ribosome biosynthesis-related proteins involved in oncogenesis. Members of one group are classified as “tumor suppressive factors” represented by RPL5 and RPL11; loss of their functions leads to cancer predisposition. RPL5 and RPL11 prevent tumorigenesis by binding to and inhibiting the MDM2 ubiquitin ligase and thereby up-regulating p53. Many other candidate tumor suppressive ribosomal/nucleolar proteins have been suggested. However, it remains to be experimentally clarified whether many of these factors can actually prevent tumorigenesis and if so, how they do so. Conversely, some ribosomal/nucleolar proteins promote tumorigenesis. For example, PICT1 binds to and anchors RPL11 in nucleoli, down-regulating p53 and promoting tumorigenesis. GRWD1 was recently identified as another such factor. When overexpressed, GRWD1 suppresses p53 and transforms normal human cells, probably by binding to RPL11 and sequestrating it from MDM2. However, other pathways may also be involved.
Introduction
Certain ribosomal or ribosome biosynthesis-related proteins (hereafter termed ribosomal/nucleolar proteins) have recently emerged as important oncogenesis-associated factors. Some act as tumor suppressors, whereas others act as tumor promoters ( and ). Tumor suppressive ribosomal/nucleolar proteins are characterized as those whose dysfunction by genetic mutations causes cancer-prone ribosomopathies such as Diamond-Blackfan anemia (DBA), 5q− syndrome, and T-cell acute lymphoblastic leukemia (T-ALL) (). Ribosomopathies are defined as disorders resulting from deficiencies in a ubiquitous and fundamental biochemical process, namely, ribosome biogenesis. Despite their common etiology, ribosomopathies show variable clinical manifestations with apparently tissue-specific phenotypes. Although the molecular mechanism underlying such heterogeneity is a very intriguing issue, it will not be discussed in this review (for reviews, seeCitation1,Citation2). Interestingly, many ribosomopathies are accompanied by a predisposition to cancer.Citation1,Citation2 It remains unclear whether other ribosomopathies are truly not associated with cancer predisposition. This is partly due to difficulties in establishing genotype-phenotype correlation in ribosomopathies with many causative genes and variable phenotypes. The existence of many causative genes also makes it difficult to experimentally explore their biological functions. Nevertheless, the ribosomal protein-MDM2-p53 axis is one well-known and well-studied pathway that may account for the cancer predisposition seen with several ribosomopathies (). For example, RPL5 (ribosomal protein L5) and RPL11, whose heterozygous mutation results in DBA, play a crucial role in this cascade.Citation1 We will concisely describe this and other possible mechanisms underlying how certain ribosomal/nucleolar proteins act as tumor suppressive factors.
Table 1. Examples of possible tumor suppressive ribosomal/nucleolar proteins.
Table 2. Examples of possible tumor-promoting ribosomal/nucleolar proteins.
Figure 1. A model for p53 regulation by ribosomal/nucleolar proteins. (A) In unstressed cells, tumor suppressive ribosomal proteins (RPs) such as RPL11 localize in the nucleolus, and MDM2 binds to and ubiquitinates p53. As a result, the cellular p53 level is kept low. (B) Nucleolar stress disrupts the nucleolus and releases the RPs into the nucleoplasm, where they interact with MDM2 and inhibit its ubiquitin ligase activity, inducing p53. (C) Other stresses such as DNA double-strand breaks and hypergrowth stimuli (e.g., expression of activated RAS) are also suggested to induce release of the RPs into the nucleoplasm, contributing to p53 induction. It is unclear whether nucleolar breakdown occurs under such conditions. (D) In cancer-prone ribosomopathy cells, the levels of the RPs are decreased by heterozygous mutations (or homozygous mutations for some factors). As a result, the RP-MDM2-p53 pathway cannot function in stressed cells, leading to tumorigenesis. (E) When overexpressed, GRWD1 binds to and inhibits RPL11, thereby down-regulating p53. As a result, tumor formation is promoted. As detailed in the text, PICT1 similarly binds to RPL11 and thereby inhibits p53.
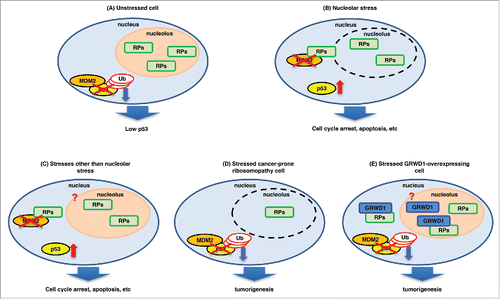
On the other hand, it was recently demonstrated that some ribosomal/nucleolar proteins conversely promote tumorigenesis by modulating the functions of tumor suppressive ribosomal proteins (). For example, PICT1 (protein interacting with C-terminus 1 of the tumor suppressor phosphatase and tensin homolog) binds to and anchors RPL11 in nucleoli, leading to p53 downregulation and promotion of tumorigenesis ().Citation3 We recently identified GRWD1 (glutamate-rich WD40 repeat-containing 1) as another such factor.Citation4 When overexpressed, GRWD1 down-regulates p53 and transforms normal human cells in combination with activated mutant KRAS and human papilloma virus E7, which inhibits RB. Consistent with these findings, high expression of GRWD1 is associated with poor prognosis of patients having certain cancers. At least one critical pathway by which GRWD1 acts is binding to RPL11 and sequestrating it from MDM2 ().Citation4 We will focus on these newly identified players that function as tumor-promoting ribosomal/nucleolar proteins. Finally, some important unclarified issues in the field that should be addressed in the future will be discussed.
Tumor suppressive ribosomal proteins that bind to MDM2 and up-regulate p53
p53 is best known for its role as a tumor suppressor.Citation1,Citation5,Citation6 Under unstressed conditions, the RING finger E3 ubiquitin ligase MDM2 negatively regulates p53 levels by promoting its polyubiquitination-mediated degradation ().Citation1,Citation5,Citation6 In addition, MDM2 can inhibit p53 transcriptional activity.Citation1,Citation5,Citation6 Other ubiquitin ligases such as Pirh2,Citation7 COP1,Citation8 and Synoviolin-19 have been also implicated in proteolytic regulation of p53. When cells encounter various stresses such as DNA damage, oncogenic activation, and nucleolar stress, p53 is activated by escaping from these ubiquitin ligases and regulates transcription of many coding and noncoding target genes. Finally, activated p53 ensures cellular homeostasis by inducing cell cycle arrest, senescence, apoptosis, changes in metabolism, and DNA repair.Citation5,Citation6,Citation10
Several lines of evidence clearly demonstrate the mechanism by which certain ribosomal proteins, such RPL5, RPL11, RPL23, RPL26, RPS3, RPS7, and RPS14, activate p53. When overexpressed, these proteins can bind to MDM2 and inhibit MDM2-mediated ubiquitination and subsequent degradation of p53 ().Citation1,Citation11-18 Among them, RPL5, RPL11, RPL26, and RPS7 are causative genes for DBA, and RPS14 is a causative gene for 5q− syndrome.Citation19-23 Under normal cell growth conditions, ribosomal proteins are assembled into nascent ribosomes. When ribosome biogenesis is perturbed, nucleolar disruption and passive diffusion of many ribosomal proteins, including RPL5 and RPL11, from the nucleolus to the nucleoplasm occurs, where they bind to MDM2 and inhibit its ubiquitination activity toward p53, leading to p53 upregulation ().Citation1,Citation10,Citation24,Citation25
In particular, RPL5 and RPL11 are regarded as crucial p53 regulators in the nucleolar stress response. RPL5 and RPL11, but not RPL23, RPL26, or RPS7, accumulate in non-ribosomal fractions upon impairment of ribosome biogenesis, where they bind to MDM2.Citation26 In addition, depletion of RPL5 or RPL11, but not of RPL23, RPL26, or RPS7, dramatically inhibits p53 induction by actinomycin D, an inducer of nucleolar stress.Citation11,Citation26-29 The importance of the RPL5/RPL11-MDM2-p53 pathway in cancer prevention is convincingly supported by an in vivo mouse model. Knock-in mice that express a MDM2 mutant, MDM2 C305F, which cannot interact with RPL5 or RPL11, exhibit impaired p53 activation upon disruption of ribosome biogenesis and promotion of tumorigenesis.Citation30 RPL5 and RPL11 also play an important role in p53 induction by DNA double-strand breaks or hypergrowth stimuli such as activated RAS or c-myc overexpression ().Citation26,Citation30-32
In addition, RPS14 may also be a critical regulator of the p53 response. It binds to MDM2 via the central acidic domain and inhibits MDM2-mediated p53 ubiquitination and degradation. Moreover, siRNA-mediated depletion of RPS14 disturbs p53 activation induced by ribosomal stress, but not by DNA damage.Citation17 While RPL26 is reported to bind to and inhibit MDM2 as described above, it may also regulate p53 mRNA translation.Citation33 In particular, RPL26 can affect p53 regulation upon irradiation.Citation33
RPL22, a lineage-specific tumor suppressive ribosomal protein
RPL22 is inactivated in less than 10% of human T-ALL cases. It is a widely expressed component of the 60S large ribosome subunit, but is not essential for core ribosome function. Moreover, germline ablation of the RPL22 gene is not lethal, and RPL22-deficient mice are of normal size, fertile, and healthy.Citation34 However, RPL22 inactivation accelerates the development of thymic lymphoma in a mouse model and also enhances the transformation of both primary and immortalized murine embryonic fibroblasts. Therefore, RPL22 may function as a haploinsufficient tumor suppressor.Citation35 As a possible mechanism underlying the transformation, RPL22 inactivation is suggested to induce the stemness factor Lin28B.Citation35
RPL22 may directly bind to p53 mRNA and down-regulate its protein synthesis in thymic double-negative cells.Citation36 RPL22 interacts with RNA targets via recognition of a stem loop structure with a G-C-U sequence at the neck of the stem.Citation37 RPL22 deficiency up-regulates p53 selectively in thymic αβ T lineage progenitors.Citation38 However, the molecular mechanism(s) underlying this selectivity and the biological relevance of such p53 regulation in tumor prevention remain to be determined.
Which candidate tumor suppressive ribosomal proteins actually suppress tumorigenesis in experimental models?
DBA is characterized by macrocytic anemia with reduced numbers of progenitors in bone marrow. In addition to anemia, DBA patients are also characterized by increased cancer susceptibility.Citation20–23,Citation39 This is caused by mutations in genes encoding components of either the small or large ribosomal subunits, including RPS7, RPS10, RPS17, RPS19, RPS24, RPS26, RPL5, RPL11, RPL26, and RPL35A.Citation1,Citation2,Citation20-23 Among the causative factors, RPL5, RPL11, RPL26, and RPS7 can bind to and inhibit MDM2, as described above. This could explain the cancer-prone phenotype of DBA caused by these factors (). It will be interesting to investigate whether other DBA factors such as RPS19, a major causative factor of DBA, affect the MDM2-p53 pathway.
Unfortunately, it remains largely unclear whether the DBA factors actually function as tumor suppressors in experimental models. Morgado-Palacin et al. generated a mouse knockout model and showed that homozygous deletion of RPL11 is lethal and that its heterozygous deletion results in chronic anemia. Interestingly, heterozygous RPL11 mutant mice are also highly susceptible to radiation-induced lymphoma.Citation32 On the other hand, the cancer-prone phenotype has not been observed in heterozygous RPS7 mutant mice.Citation40
The RPS19 gene is most frequently mutated (about 25%) in DBA.Citation20,Citation41,Citation42 It remains unclear whether RPS19 can affect the MDM2-p53 pathway. In transgenic mice expressing shRNA against RPS19 under the control of an inducible promotor, RPS19 deficiency induces bone marrow failure, which is rescued by p53 inactivation.Citation43 On the other hand, Matsson et al. reported that embryos with heterozygous RPS19 deletion develop normally, showing no feature of human DBA later in life.Citation44 Thus, it remains unclear whether RPS19 plays a role in tumor suppression in experimental models.
5q− syndrome is caused by a somatically acquired deletion in the long arm of chromosome 5 and is characterized by macrocytic anemia with decreased erythroid progenitors in bone marrow and an increased risk of developing acute myeloid leukemia, similar to DBA.Citation45 RPS14 is thought to be a causative agent of 5q− syndrome.Citation19,Citation45-49 As described above, RPS14 can bind to and inhibit MDM2.Citation17 However, RPS14 haploinsufficiency induces macrocytic anemia but not cancer predisposition in mouse models.Citation50,Citation51
The oncogenic ribosomal/nucleolar protein PICT1 down-regulates RPL11-mediated p53 activation
A recent study suggests that PICT1, also known as GLTSCR2, down-regulates RPL11-mediated p53 activation.Citation3 Because PICT1 stabilizes phosphatase and tensin homolog, a tumor suppressor,Citation52 and its low expression in diffuse glioma and ovarian cancer is correlated with poor prognosis,Citation52-54 it was originally proposed to be a tumor suppressor. In addition, PICT1 overexpression induces apoptosis in cultured glioma cells. On the other hand, individuals with gastric, colorectal, esophageal, and hepatocellular carcinoma, low PICT1 expression, and wild-type p53 have a better prognosis.Citation3,Citation55,Citation56 Moreover, patients having oligodendrogliomas with PICT1 haploinsufficiency have a better prognosis than other patients.Citation57–59 In addition, high expression of PICT1 is associated with poor prognosis in non-small cell lung cancer.Citation60 These findings suggest that PICT1 rather functions as an oncogene.
Sasaki et al. showed that PICT1 inhibits p53 via the RPL11-MDM2-p53 pathway (). In normal cells, PICT1 binds to and retains RPL11 in the nucleolus, and its deficiency induces RPL11 translocation to the nucleoplasm. The released RPL11 then binds to and inhibits MDM2, leading to p53 activation.Citation3,Citation55,Citation61 A recent study suggests that PICT1 directly binds to 5S rRNA in nucleoli and plays an essential role in 5S RNP (the complex of 5S rRNA, RPL5, and RPL11) integration into ribosomes.Citation62 Furthermore, Maehama et al. reported that nucleolar stress induces proteasome-mediated PICT1 degradation, probably to allow RPL11 to bind to MDM2.Citation63 Although detailed molecular mechanism(s) remains elusive, the PICT1 degradation appears to occur in a polyubiquitination-independent manner.Citation63 In addition, PICT1 is also suggested to be phosphorylated by ATM in response to DNA damage and then degraded. Because PICT1 downregulation leads to p53 activation as described above, this pathway could contribute to the cellular DNA damage response.Citation64 Taken together, these results demonstrate that PICT1 is a negative regulator of the p53 response and a potential oncogene. However, it is still unclear whether PICT1 overexpression actually transforms cells in any experimental models.
The novel oncogenic ribosomal/nucleolar protein GRWD1 down-regulates p53 and promotes tumorigenesis
GRWD1, a WD40 protein that is highly conserved among eukaryotes, has been functionally implicated in ribosome biogenesis.Citation65–67 We identified GRWD1 as a novel Cdt1-binding proteinCitation68 and later found that it possesses histone-binding and nucleosome assembly activities and promotes MCM loading, probably by maintaining chromatin openness at replication origins.Citation69 Cdt1 and MCM are essential proteins that form pre-replication complexes required for DNA replication initiation.Citation70 Furthermore, we recently reported that GRWD1 may promote chromatin fluidity by influencing nucleosome structures, e.g., by transient eviction of H2A-H2B.Citation71 On the other hand, GRWD1 is one of the DDB1-interacting WD40 proteins, which are predicted to function as substrate receptors of the Cul4-DDB1 ubiquitin ligase.Citation72,Citation73
Recently, we demonstrated that GRWD1 is a novel oncogene ().Citation4 We previously found that GRWD1 is overexpressed in cancer cells.Citation69 In a recent paper, we further found that GRWD1 depletion enhances p53 stabilization induced by various stresses, and its overexpression conversely down-regulates p53. Interestingly, in a soft agar colony formation assay and a tumorigenicity assay in nude mice, GRWD1 overexpression, in combination with E7 and activated KRAS, promotes oncogenic transformation of human normal fibroblasts, probably by repressing p53. Furthermore, high expression of GRWD1 is highly associated with poor prognosis in lower-grade brain glioma patients with wild-type p53, but not in those with mutated p53.Citation4 Together with the other functions described above, GRWD1 is a multifunctional protein involved in multiple cellular regulatory pathways, particularly those associated with cell growth control.
GRWD1 down-regulates p53 by binding to RPL11 and sequestrating it from MDM2
How does GRWD1 inhibit p53 and promote tumorigenesis? We identified RPL11 as a novel GRWD1 interactor and further found that GRWD1 interferes with the p53 activation pathway via the RPL11-MDM2 axis by interacting with RPL11 and sequestering it from MDM2 ().Citation4 As described above, GRWD1 overexpression can confer a tumorigenic capacity in normal human cells. Importantly, this activity is lost with a GRWD1 mutant that cannot interact with RPL11, suggesting that the oncogenic activity of GRWD1 is mediated, at least partly, by its binding to and inhibition of RPL11.Citation4 Therefore, it will be possible to recapitulate cellular transformation by depleting tumor suppressive ribosomal proteins such as RPL11 (in combination with E7 and activated KRAS) using this experimental system.
Concluding remarks and perspective
As summarized above, increasing attention has been paid to many ribosomal/nucleolar proteins as new regulators of oncogenesis. For tumor suppressive ribosomal proteins whose mutations lead to cancer-prone disorders, it is clear that p53 is a critical target molecule. However, it is not yet completely understood how these various factors actually prevent tumorigenesis. For example, it remains unclear how RPS19, a major DBA factor, functions in cancer prevention. A more critical problem is that the tumor suppressive activities of most factors have not been experimentally observed, as discussed above. Extensive generation of the relevant knockout mice would be useful, but is too time-consuming and expensive. In addition, despite the apparent p53 inhibitory activity of RPS14, a causative factor of cancer-prone 5q− syndrome, its haploinsufficiency has not recapitulated cancer predisposition in mouse models to date. In this regard, it will be intriguing to test whether (partial) inhibition of these factors in combination with E7 and activated KRAS can transform normal human cells. This relatively simple assay would be applicable to comprehensive investigation of candidate tumor suppressive (or tumor-promoting) ribosomal/nucleolar proteins.
On the other hand, PICT1 and GRWD1 have emerged as novel oncogenic ribosomal/nucleolar proteins. The oncogenic potential of both factors may be mainly mediated by binding to the tumor suppressive protein RPL11 and sequestering it from MDM2. Clinical data indicate that high expression of these factors is associated with poor prognosis in patients with wild-type p53, at least in certain cancer types. However, the possible relationship between PICT1 and GRWD1 has not been uncovered and should be investigated in the future.
Although the oncogenic ability of GRWD1 is mainly executed by its binding to RPL11, the possibility that GRWD1 overexpression up-regulates ribosome biogenesis and/or DNA replication, contributing to its oncogenic activity, cannot be excluded. Finally, given that GRWD1 also interacts with other growth regulatory factors including RPL23 (Citationref. 4 and our unpublished data), it remains possible that another pathway(s) also contributes to the oncogenic activity of GRWD1. It will also be interesting to address this issue in the future.
Disclosure of potential conflicts of interest
The authors declare no conflicts of interest in relation to this article.
Acknowledgment
We thank all members of our research group for helpful discussions and critical reading of the manuscript.
Funding
This work was supported in part by grants to Nozomi Sugimoto and Masatoshi Fujita from the Ministry of Education, Science, Sports, Technology and Culture of Japan.
References
- Bursac S, Brdovcak MC, Donati G, Volarevic S. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim Biophys Acta 2014; 1842:817-30; PMID:24514102; https://doi.org/10.1016/j.bbadis.2013.08.014
- Armistead J, Triggs-Raine B. Diverse diseases from a ubiquitous process: The ribosomopathy paradox. FEBS Lett 2014; 588:1491-500; PMID:24657617; https://doi.org/10.1016/j.febslet.2014.03.024
- Sasaki M, Kawahara K, Nishio M, Mimori K, Kogo R, Hamada K, Itoh B, Wang J, Komatsu Y, Yang YR, et al. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med 2011; 17:944-51; PMID:21804542; https://doi.org/10.1038/nm.2392
- Kayama K, Watanabe S, Takafuji T, Tsuji T, Hironaka K, Matsumoto M, Nakayama KI, Enari M, Kohno T, Shiraishi K, et al. GRWD1 negatively regulates p53 via the RPL11-MDM2 pathway and promotes tumorigenesis. EMBO Rep 2017; 18:123-37; PMID:27856536; https://doi.org/10.15252/embr.201642444
- Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell 2009; 137:413-31; PMID:19410540; https://doi.org/10.1016/j.cell.2009.04.037
- Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 2009; 1:a001883; PMID:20066118; https://doi.org/10.1101/cshperspect.a001883
- Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003; 112:779-91; PMID:12654245; https://doi.org/10.1016/S0092-8674(03)00193-4
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004; 429:86-92; PMID:15103385; https://doi.org/10.1038/nature02514
- Yamasaki S, Yagishita N, Sasaki T, Nakazawa M, Kato Y, Yamadera T, Bae E, Toriyama S, Ikeda R, Zhang L, et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘synoviolin’. EMBO J 2007; 26:113-22; PMID:17170702; https://doi.org/10.1038/sj.emboj.7601490
- Zhang Y, Lu H. Signaling to p53: Ribosomal proteins find their way. Cancer Cell 2009; 16:369-77; PMID:19878869; https://doi.org/10.1016/j.ccr.2009.09.024
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem 2004; 279:44475-82; PMID:15308643; https://doi.org/10.1074/jbc.M403722200
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 2004; 24:7654-68; PMID:15314173; https://doi.org/10.1128/MCB.24.17.7654-7668.2004
- Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol 2004; 24:7669-80; PMID:15314174; https://doi.org/10.1128/MCB.24.17.7669-7680.2004
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell 2008; 32:180-9; PMID:18951086; https://doi.org/10.1016/j.molcel.2008.08.031
- Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair (Amst) 2009; 8:1215-24; PMID:19656744; https://doi.org/10.1016/j.dnarep.2009.07.003
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 2003; 23:8902-12; PMID:14612427; https://doi.org/10.1128/MCB.23.23.8902-8912.2003
- Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene 2013; 32:388-96; PMID:22391559; https://doi.org/10.1038/onc.2012.63
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 2009; 35:316-26; PMID:19683495; https://doi.org/10.1016/j.molcel.2009.07.014
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 2008; 451:335-9; PMID:18202658; https://doi.org/10.1038/nature06494
- Farrar JE, Dahl N. Untangling the phenotypic heterogeneity of diamond blackfan anemia. Semin Hematol 2011; 48:124-35; PMID:21435509; https://doi.org/10.1053/j.seminhematol.2011.02.003
- Gazda HT, Preti M, Sheen MR, O'Donohue MF, Vlachos A, Davies SM, Kattamis A, Doherty L, Landowski M, Buros C, et al. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-blackfan anemia. Hum Mutat 2012; 33:1037-44; PMID:22431104; https://doi.org/10.1002/humu.22081
- Horos R, Ijspeert H, Pospisilova D, Sendtner R, Andrieu-Soler C, Taskesen E, Nieradka A, Cmejla R, Sendtner M, Touw IP, et al. Ribosomal deficiencies in diamond-blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood 2012; 119:262-72; PMID:22058113; https://doi.org/10.1182/blood-2011-06-358200
- Boria I, Garelli E, Gazda HT, Aspesi A, Quarello P, Pavesi E, Ferrante D, Meerpohl JJ, Kartal M, Da Costa L, et al. The ribosomal basis of diamond-blackfan anemia: Mutation and database update. Hum Mutat 2010; 31:1269-79; PMID:20960466; https://doi.org/10.1002/humu.21383
- Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: Probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 2010; 29:4253-60; PMID:20498634; https://doi.org/10.1038/onc.2010.189
- Zhou X, Liao JM, Liao WJ, Lu H. Scission of the p53-MDM2 loop by ribosomal proteins. Genes Cancer 2012; 3:298-310; PMID:23150763; https://doi.org/10.1177/1947601912455200
- Bursac S, Brdovcak MC, Pfannkuchen M, Orsolic I, Golomb L, Zhu Y, Katz C, Daftuar L, Grabusic K, Vukelic I, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci U S A 2012; 109:20467-72; PMID:23169665; https://doi.org/10.1073/pnas.1218535109
- Daftuar L, Zhu Y, Jacq X, Prives C. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS One 2013; 8:e68667; PMID:23874713; https://doi.org/10.1371/journal.pone.0068667
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 2003; 3:577-87; PMID:12842086; https://doi.org/10.1016/S1535-6108(03)00134-X
- Fumagalli S, Ivanenkov VV, Teng T, Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev 2012; 26:1028-40; PMID:22588717; https://doi.org/10.1101/gad.189951.112
- Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, Zhang Y. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell 2010; 18:231-43; PMID:20832751; https://doi.org/10.1016/j.ccr.2010.08.007
- Nishimura K, Kumazawa T, Kuroda T, Katagiri N, Tsuchiya M, Goto N, Furumai R, Murayama A, Yanagisawa J, Kimura K. Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep 2015; 10:1310-23; PMID:25732822; https://doi.org/10.1016/j.celrep.2015.01.055
- Morgado-Palacin L, Varetti G, Llanos S, Gomez-Lopez G, Martinez D, Serrano M. Partial loss of Rpl11 in adult mice recapitulates diamond-blackfan anemia and promotes lymphomagenesis. Cell Rep 2015; 13:712-22; PMID:26489471; https://doi.org/10.1016/j.celrep.2015.09.038
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 2005; 123:49-63; PMID:16213212; https://doi.org/10.1016/j.cell.2005.07.034
- Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity 2007; 26:759-72; PMID:17555992; https://doi.org/10.1016/j.immuni.2007.04.012
- Rao S, Lee SY, Gutierrez A, Perrigoue J, Thapa RJ, Tu Z, Jeffers JR, Rhodes M, Anderson S, Oravecz T, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 2012; 120:3764-73; PMID:22976955; https://doi.org/10.1182/blood-2012-03-415349
- Rashkovan M, Vadnais C, Ross J, Gigoux M, Suh WK, Gu W, Kosan C, Moroy T. Miz-1 regulates translation of Trp53 via ribosomal protein L22 in cells undergoing V(D)J recombination. Proc Natl Acad Sci U S A 2014; 111:E5411-5419; PMID:25468973; https://doi.org/10.1073/pnas.1412107111
- Dobbelstein M, Shenk T. In vitro selection of RNA ligands for the ribosomal L22 protein associated with epstein-barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J Virol 1995; 69:8027-34; PMID:7494316
- Solanki NR, Stadanlick JE, Zhang Y, Duc AC, Lee SY, Lauritsen JP, Zhang Z, Wiest DL. Rpl22 loss selectively impairs alphabeta T cell development by dysregulating endoplasmic reticulum stress signaling. J Immunol 2016; 197:2280-9; PMID:27489283; https://doi.org/10.4049/jimmunol.1600815
- Ruggero D, Shimamura A. Marrow failure: A window into ribosome biology. Blood 2014; 124:2784-92; PMID:25237201; https://doi.org/10.1182/blood-2014-04-526301
- Watkins-Chow DE, Cooke J, Pidsley R, Edwards A, Slotkin R, Leeds KE, Mullen R, Baxter LL, Campbell TG, Salzer MC, et al. Mutation of the diamond-blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet 2013; 9:e1003094; PMID:23382688; https://doi.org/10.1371/journal.pgen.1003094
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in diamond-blackfan anaemia. Nat Genet 1999; 21:169-75; PMID:9988267; https://doi.org/10.1038/5951
- Willig TN, Draptchinskaia N, Dianzani I, Ball S, Niemeyer C, Ramenghi U, Orfali K, Gustavsson P, Garelli E, Brusco A, et al. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: Wide variations in phenotypic expression. Blood 1999; 94:4294-306; PMID:10590074
- Jaako P, Flygare J, Olsson K, Quere R, Ehinger M, Henson A, Ellis S, Schambach A, Baum C, Richter J, et al. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with diamond-blackfan anemia. Blood 2011; 118:6087-96; PMID:21989989; https://doi.org/10.1182/blood-2011-08-371963
- Matsson H, Davey EJ, Draptchinskaia N, Hamaguchi I, Ooka A, Leveen P, Forsberg E, Karlsson S, Dahl N. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol 2004; 24:4032-7; PMID:15082795; https://doi.org/10.1128/MCB.24.9.4032-4037.2004
- Narla A, Ebert BL. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010; 115:3196-205; PMID:20194897; https://doi.org/10.1182/blood-2009-10-178129
- Barlow JL, Drynan LF, Trim NL, Erber WN, Warren AJ, McKenzie AN. New insights into 5q- syndrome as a ribosomopathy. Cell Cycle 2010; 9:4286-93; PMID:20980806; https://doi.org/10.4161/cc.9.21.13742
- Boultwood J. The role of haploinsufficiency of RPS14 and p53 activation in the molecular pathogenesis of the 5q- syndrome. Pediatr Rep 2011; 3 Suppl 2:e10; PMID:22053272; https://doi.org/10.4081/pr.2011.s2.e10
- Boultwood J, Pellagatti A, Cattan H, Lawrie CH, Giagounidis A, Malcovati L, Della Porta MG, Jadersten M, Killick S, Fidler C, et al. Gene expression profiling of CD34+ cells in patients with the 5q- syndrome. Br J Haematol 2007; 139:578-89; PMID:17916100; https://doi.org/10.1111/j.1365-2141.2007.06833.x
- Wu L, Li X, Xu F, Zhang Z, Chang C, He Q. Low RPS14 expression in MDS without 5q - aberration confers higher apoptosis rate of nucleated erythrocytes and predicts prolonged survival and possible response to lenalidomide in lower risk non-5q- patients. Eur J Haematol 2013; 90:486-93; PMID:23506134; https://doi.org/10.1111/ejh.12105
- Schneider RK, Schenone M, Ferreira MV, Kramann R, Joyce CE, Hartigan C, Beier F, Brummendorf TH, Germing U, Platzbecker U, et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat Med 2016; 22:288-97; PMID:26878232; https://doi.org/10.1038/nm.4047
- Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med 2010; 16:59-66; PMID:19966810; https://doi.org/10.1038/nm.2063
- Yim JH, Kim YJ, Ko JH, Cho YE, Kim SM, Kim JY, Lee S, Park JH. The putative tumor suppressor gene GLTSCR2 induces PTEN-modulated cell death. Cell Death Differ 2007; 14:1872-9; PMID:17657248; https://doi.org/10.1038/sj.cdd.4402204
- Merritt MA, Parsons PG, Newton TR, Martyn AC, Webb PM, Green AC, Papadimos DJ, Boyle GM. Expression profiling identifies genes involved in neoplastic transformation of serous ovarian cancer. BMC Cancer 2009; 9:378, 2407-9-378; PMID:19849863; https://doi.org/10.1186/1471-2407-9-378
- Kim YJ, Cho YE, Kim YW, Kim JY, Lee S, Park JH. Suppression of putative tumour suppressor gene GLTSCR2 expression in human glioblastomas. J Pathol 2008; 216:218-24; PMID:18729076; https://doi.org/10.1002/path.2401
- Uchi R, Kogo R, Kawahara K, Sudo T, Yokobori T, Eguchi H, Sugimachi K, Maehama T, Mori M, Suzuki A, et al. PICT1 regulates TP53 via RPL11 and is involved in gastric cancer progression. Br J Cancer 2013; 109:2199-206; PMID:24045667; https://doi.org/10.1038/bjc.2013.561
- Ishibashi M, Kogo R, Shibata K, Ueo H, Uchi R, Matsumura T, Takano Y, Sawada G, Takahashi Y, Mima K, et al. Clinical significance of PICT1 in patients of hepatocellular carcinoma with wild-type TP53. Ann Surg Oncol 2013; 20 Suppl 3:S537-544; PMID:23532381; https://doi.org/10.1245/s10434-013-2958-x
- Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G, Hosek SM, Kimmel D, O'Fallon J, Yates A, et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene 1999; 18:4144-52; PMID:10435596; https://doi.org/10.1038/sj.onc.1202759
- Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 1998; 90:1473-9; PMID:9776413
- Mariani L, Deiana G, Vassella E, Fathi AR, Murtin C, Arnold M, Vajtai I, Weis J, Siegenthaler P, Schobesberger M, et al. Loss of heterozygosity 1p36 and 19q13 is a prognostic factor for overall survival in patients with diffuse WHO grade 2 gliomas treated without chemotherapy. J Clin Oncol 2006; 24:4758-63; PMID:16966689; https://doi.org/10.1200/JCO.2006.05.9238
- Okamura K, Takayama K, Kawahara K, Harada T, Nishio M, Otsubo K, Ijichi K, Kohno M, Iwama E, Fujii A, et al. PICT1 expression is a poor prognostic factor in non-small cell lung cancer. Oncoscience 2014; 1:375-82; PMID:25594032; https://doi.org/10.18632/oncoscience.43
- Suzuki A, Kogo R, Kawahara K, Sasaki M, Nishio M, Maehama T, Sasaki T, Mimori K, Mori M. A new PICTure of nucleolar stress. Cancer Sci 2012; 103:632-7; PMID:22320853; https://doi.org/10.1111/j.1349-7006.2012.02219.x
- Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 2013; 5:237-47; PMID:24120868; https://doi.org/10.1016/j.celrep.2013.08.049
- Maehama T, Kawahara K, Nishio M, Suzuki A, Hanada K. Nucleolar stress induces ubiquitination-independent proteasomal degradation of PICT1 protein. J Biol Chem 2014; 289:20802-12; PMID:24923447; https://doi.org/10.1074/jbc.M114.571893
- Chen H, Han L, Tsai H, Wang Z, Wu Y, Duo Y, Cao W, Chen L, Tan Z, Xu N, et al. PICT-1 is a key nucleolar sensor in DNA damage response signaling that regulates apoptosis through the RPL11-MDM2-p53 pathway. Oncotarget 2016; 7:83241-57; PMID:27829214; https://doi.org/10.18632/oncotarget.13082
- Gratenstein K, Heggestad AD, Fortun J, Notterpek L, Pestov DG, Fletcher BS. The WD-repeat protein GRWD1: Potential roles in myeloid differentiation and ribosome biogenesis. Genomics 2005; 85:762-73; PMID:15885502; https://doi.org/10.1016/j.ygeno.2005.02.010
- Iouk TL, Aitchison JD, Maguire S, Wozniak RW. Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis. Mol Cell Biol 2001; 21:1260-71; https://doi.org/10.1128/MCB.21.4.1260-1271.2001
- Schaper S, Fromont-Racine M, Linder P, de la Cruz J, Namane A, Yaniv M. A yeast homolog of chromatin assembly factor 1 is involved in early ribosome assembly. Curr Biol 2001; 11:1885-90; https://doi.org/10.1016/s0960-9822(01)00584-x
- Sugimoto N, Kitabayashi I, Osano S, Tatsumi Y, Yugawa T, Narisawa-Saito M, Matsukage A, Kiyono T, Fujita M. Identification of novel human Cdt1-binding proteins by a proteomics approach: Proteolytic regulation by APC/CCdh1. Mol Biol Cell 2008; 19:1007-21; PMID:18162579; https://doi.org/10.1091/mbc.E07-09-0859
- Sugimoto N, Maehara K, Yoshida K, Yasukouchi S, Osano S, Watanabe S, Aizawa M, Yugawa T, Kiyono T, Kurumizaka H, et al. Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res 2015; 43:5898-911; PMID:25990725; https://doi.org/10.1093/nar/gkv509
- Higa M, Fujita M, Yoshida K. DNA replication origins and fork progression at mammalian telomeres. Genes (Basel) 2017; 8:112; PMID:28350373; https://doi.org/10.3390/genes8040112.
- Aizawa M, Sugimoto N, Watanabe S, Yoshida K, Fujita M. Nucleosome assembly and disassembly activity of GRWD1, a novel Cdt1-binding protein that promotes pre-replication complex formation. Biochim Biophys Acta 2016; 1863:2739-48; PMID:27552915; https://doi.org/10.1016/j.bbamcr.2016.08.008
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev 2006; 20:2949-54; https://doi.org/10.1101/gad.1483206
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 2006; 8:1277-83; https://doi.org/10.1038/ncb1490
- Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, Bessler M, Mason PJ. Cells depleted for RPS19, a protein associated with diamond blackfan anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis 2007; 39:35-43; PMID:17376718; https://doi.org/10.1016/j.bcmd.2007.02.001
- Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. Impaired ribosome biogenesis in diamond-blackfan anemia. Blood 2007; 109:1275-83; PMID:17053056; https://doi.org/10.1182/blood-2006-07-038372
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet 2013; 45:186-90; PMID:23263491; https://doi.org/10.1038/ng.2508
