ABSTRACT
Profilin-1 (Pfn1) is an important actin-regulatory protein that is downregulated in human breast cancer and when forcibly elevated, it suppresses the tumor-initiating ability of triple-negative breast cancer cells. In this study, we demonstrate that Pfn1 overexpression reduces the stem-like phenotype (a key biologic feature associated with higher tumor-initiating potential) of MDA-MB-231 (MDA-231) triple-negative breast cancer cells. Interestingly, the stem-like trait of MDA-231 cells is also attenuated upon depletion of Pfn1. A comparison of cancer stem cell gene (CSC) gene expression signatures between depleted and elevated conditions of Pfn1 further suggest that Pfn1 may be somehow involved in regulating the expression of a few CSC-related genes including MUC1, STAT3, FZD7, and ITGB1. Consistent with the reduced stem-like phenotype associated with loss-of-function of Pfn1, xenograft studies showed lower tumor-initiating frequency of Pfn1-depleted MDA-231 cells compared to their control counterparts. In MMTV:PyMT mouse model, homozygous but not heterozygous deletion of Pfn1 gene leads to severe genetic mosaicism and positive selection of Pfn1-proficient tumor cells further supporting the contention that a complete lack of Pfn1 is likely not conducive for efficient tumor initiation capability of breast cancer cells. In summary, these findings suggest that the maintenance of optimal stemness and tumor-initiating ability of breast cancer cells requires a balanced expression of Pfn1.
Introduction
Disrupted actin cytoskeleton is a hallmark of cancer cells. Lower filamentous actin (F-actin) density correlates with higher grade of breast cancer, suggesting that cytoskeletal disruption likely contributes to aggressiveness of breast cancer cells.Citation1 Deregulated expression and/or activity of various actin-binding proteins and their upstream regulators are responsible for altered cytoskeleton in various cancer cells. Along this line, profilin-1 (Pfn1), the ubiquitously expressed isoform of Pfn family of actin-binding proteins that also interacts with a diverse range of protein ligands besides actin and even certain membrane lipids, is downregulated but not totally depleted in human breast cancer.Citation2,3 There is experimental evidence for Pfn1's ability to strongly suppress tumor-initiating ability of triple-negative breast cancer cell lines (CAL51, MDA-MB-231 (referred to as MDA-231 from hereon)) when forcibly elevated even to a moderate extent.Citation2,Citation4 Pfn1's tumor-suppressive action in breast cancer cells requires its interaction with actin.Citation5However, the molecular mediators and pathways responsible for Pfn1's tumor-suppressive action in breast cancer cells have not been clearly identified. Furthermore, whether Pfn1 downregulation has any impact on tumor-initiating ability of breast cancer cells has not been investigated.
Breast cancer is now widely recognized as a heterogeneous disease that has cellular hierarchy similar to the normal tissue stem cell hierarchy. Hierarchically organized tumor tissue contains a small subpopulation of cancer cells that exhibits the traits of stem cells with self-renewing property and ability for sustenance of long-term clonal maintenance in the tumor. The pool of breast cancer cells that have stem- or progenitor-like characteristics have much higher tumor-initiating potential than those in non-stem cell state (constitute the bulk of the tumor), and are generally thought to be the true tumor-initiating cells in a neoplasm.Citation6
A few reports have indicated involvement of Pfn in regulating stem cell fate. For example, Pfn1 has been shown to be essential for maintenance of hematopoeitic stem cells (HSC).Citation7 Specifically, this study showed that Pfn1 depletion compromises HSC retention via promoting their apoptosis and induction of cell-cycle quiescence. Another study showed that Pfn2 (the minor isoform of Pfn) promotes stemness of colorectal cancer cells.Citation8 Although these studies support the ability of both Pfn isoforms to regulate stem cell fate, since overexpression of Pfn1 alone is sufficient to suppress tumorigenic phenotype of triple-negative breast cancer cells,Citation2,Citation4 in this study we explored the effects of selective perturbations of Pfn1 on CSC-related gene expression and stem-cell-like traits of breast cancer cells. We show that either elevation or depletion of Pfn1 attenuates stem-like phenotype of breast cancer cells. Correlated with reduced stemness of breast cancer cells upon Pfn1 depletion, we further showed that loss-of-expression of Pfn1 also reduces the tumor-initiating potential of breast cancer cells. In light of previous findings of Pfn1-induced suppression of tumorigenicity of breast cancer cells,Citation2,Citation4 these data suggest that a balanced level of Pfn1 is required for maintenance of optimal stemness and tumor-initiating potential of breast cancer cells.
Results
A balanced level of Pfn1 promotes stemness of MDA-231 breast cancer cells
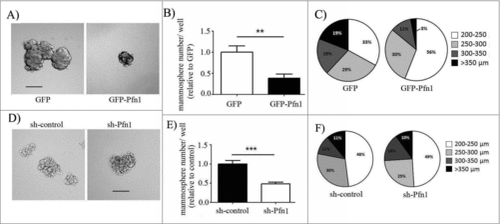
We previously reported that overexpression of GFP-Pfn1 completely suppresses the tumorigenic ability of orthotopically xenografted MDA-231 cells in nude mice. Since breast cancer stem and progenitor cells have the ability to form mammospheres on non-adherent substrates, to assess the effect of Pfn1 overexpression on stemness of MDA-231 cells, we performed mammosphere assays with sublines of MDA-231 cells stably expressing either GFP (control) or GFP-Pfn1 (this cell line has a nearly 2-fold overexpression of Pfn1Citation9). Note that MDA-231 cells have ∼100-fold lower Pfn2 level (in a sub-micromolar range of concentration) than Pfn1 (in tens of micromolar range of concentration) as previously estimated by another groupCitation1 and furthermore, stable overexpression of GFP-Pfn1 in this cell line does not lead to any appreciable change in Pfn2 expression (Fig S1). We found that GFP-Pfn1 overexpressing MDA-231 cells formed significantly fewer mammospheres than control GFP cells, suggesting that Pfn1 elevation attenuates the stemness-enriched pool of MDA-231 cells (). Overexpression of Pfn1 also limited the growth of mammospheres as evident from significantly higher fraction of mammospheres in smaller sized groups (). This may suggest that self-renewal capacity of breast cancer stem/progenitor cells is also suppressed by Pfn1 overexpression. As a complementary experiment, we next investigated the effect of Pfn1 depletion on mammosphere forming ability of MDA-231 cells using our previously generated sublines of MDA-231 cells stably expressing either Pfn1-shRNA (which had >90% knockdown of Pfn1 expression) or control luciferase shRNA.Citation3 Note that Pfn2 expression is more or less unaffected upon stable knockdown of Pfn1 expression in MDA-231 cells (Fig. S1) and this is similar to our observation in transient knockdown setting.Citation10 Interestingly, similar to the effect of Pfn1 overexpression, there was a marked reduction in the number of MDA-231 mammospheres in Pfn1 knockdown compared to the control culture (), suggesting that loss of Pfn1 expression also attenuates the stemness-enriched pool of MDA-231 cells. However, unlike our observation in the overexpression setting, the size distributors of mammospheres between control and Pfn1 knockdown groups were comparable (). One possible interpretation of this finding could be that Pfn1 depletion only reduces the stemness-enriched pool but does not impact the sustained proliferation of stem/progenitor-like cells. Overall, these data suggest that a balanced level of Pfn1 is required for maintenance of optimal stemness of breast cancer cells.
Figure 1. Effects of perturbations of Pfn1 on mammosphere forming ability of MDA-231 cells. (A, D) Representative images of mammosphere formation by GFP control vs GFP-Pfn1 overexpressers (A) and control- vs Pfn1 shRNA expressers (D) of MDA-231 cells (scale bars – 200 μm). (B, C, E, F) Bar graphs (B, E) and pie charts (C, F) summarize the number and the size distribution of MDA-231 mammospheres, respectively, in overexpression and knockdown settings of Pfn1. Data summarized from 3 independent experiments and values are presented as mean ± SD; **p < 0.01; ***:p < 0.001).
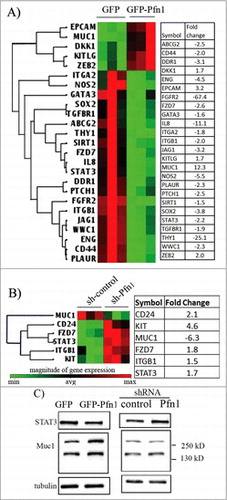
We next profiled expression of a panel of 84 CSC-related genes in MDA-231 cells under both overexpression and knockdown settings of Pfn1 by qPCR array. Filtering the genes using a 1.5-fold cut-off revealed a dramatic alteration in the expression profile of stem-cell associated genes upon Pfn1 overexpression as evident from differential expression of 25 out of a total of 84 profiled genes with 80% of those differentially expressed genes found to be downregulated in Pfn1 overexpressing cells (). These included genes that a) are required for maintenance and self-renewal of stem cells (e.g. SOX2- reduced by 3.8-fold; FGFR2- reduced by 67-fold; CD44 – reduced by 2-fold; ITGB1 – reduced by 2-fold),Citation11-14 b) expand the pool of breast CSC and increase their self-renewal (e.g., IL8 - reduced by 11-fold; STAT3 – reduced by 2-fold),Citation15,Citation16 and c) enhance CSC activities of breast cancer cells indirectly through facilitating their interactions with tumor-associated macrophages (e.g., Thy1 - reduced by 11-fold).Citation17 Compared to the effect of Pfn1 overexpression, the CSC gene expression signature of MDA-231 was much less perturbed upon Pfn1 depletion. We found a total of 6 differentially expressed genes (MUC1, CD24, Kit, FZD7, ITGB1 and STAT3) associated with Pfn1 knockdown (). Among these 6 genes, MUC1 (a gene that is overexpressed in breast CSC and promotes self-renewal capacity of breast CSCCitation18,Citation19) showed the most prominent change (> 6-fold downregulation) in expression as a result of Pfn1 knockdown. Breast CSC are also characterized by high CD44 and low CD24 expression.Citation11 Therefore, a 2-fold increase in CD24 expression also appears to be consistent with reduced stem-like phenotype of breast cancer cells upon Pfn1 depletion. Interestingly, the remaining 3 CSC-promoting genes including Kit1, FZD7 (frizzled 7), ITGB1 and STAT3 were found to be elevated although other than Kit1, the fold-change of remaining genes was modest (< 2-fold). Note that 4 out 6 genes that were differentially expressed upon knockdown of Pfn1 (MUC1, STAT3, FZD7, ITGB1) also showed a reverse trend in the expression upon overexpression of Pfn1 which suggests that Pfn1 may be somehow involved in regulating the pathways controlling the expression of at least some of this small subset of genes. As a further proof-of- principle of Pfn1s link to the regulation of some of these genes at the protein level, we performed immunoblot analyses of STAT3 and MUC1 in MDA-231 cells under depleted and overexpressed conditions of Pfn1 (). Consistent with the qPCR array results, we found a trend of increased and decreased STAT3 expression in response to knockdown and overexpression of Pfn1, respectively. Similarly, MUC1 expression was increased upon overexpression of Pfn1 as expected from the qPCR results although MUC1 increase at the protein level (∼2-fold) did not quantitatively correlate with the fold-change in transcript level (∼12-fold). We were also unable to detect any significant difference in the MUC1 protein level between control and Pfn1 knockdown cells (the plausible reasons of discrepancies between qPCR and immunoblot readouts are discussed later).
Figure 2. Effects of perturbations of Pfn1 on CSC gene expression signature in MDA-231 cells. (A-B) Cluster diagrams depicting differentially expressed (fold-change cut-off = 1.5; p-value < 0.05) CSC-related genes in response to overexpression (A) and knockdown (B) of Pfn1 expression in MDA-231 cells, based on the readouts of qPCR array. The tables alongside display the fold-change of each differentially expressed gene (data summarized from 3 independent experiments). (C) Immunoblot analyses of STAT3 and MUC1 expression from whole cell lysates of control vs Pfn1 knockdown and GFP vs GFP-Pfn1 overexpressing MDA-231 cells (tubulin blot serves as the loading control). The multiple bands of MUC1 represent proteins with different degrees of glycosylation.
Loss of Pfn1 expression negatively impacts tumor-initiating ability of breast cancer cells
Given our finding that depletion of Pfn1 also attenuates the stemness-enriched pool of MDA-231 cells similar to the effect of Pfn1 elevation, we next asked whether loss-of-expression of Pfn1 has any negative impact on the tumor-initiating ability of breast cancer cells. To address this, we performed orthotopic xenograft experiments with control vs Pfn1-shRNA expressing MDA-231 cells at 2 different inoculum conditions (1 and 2 million). For each of these inoculum conditions, tumor incidence frequency was scored and compared between the 2 groups of animals following monitoring of the animals up to a period of 6 weeks, the results of which are shown in . Control shRNA transfectants were able to elicit primary tumors in 100% of animals for either of the inoculum conditions (8 out of 8 and 6 out of 6 for 1 million and 2 million cell injections, respectively). The primary tumor-initiating frequency of Pfn1-shRNA cells (50% (4 out 8) and 82% (5 out of 6) for 1- and 2-million cell injections, respectively) were lower than the control cells, with the difference being more pronounced at the lower inoculum conditions, suggesting that Pfn1 downregulation reduces tumor-initiating ability of breast cancer cells.
Although widely used, xenograft experiments are performed in immunodeficient background and involve experimental implantation of a large number of tumor cells that fails to recapitulate the initiation of human disease. Therefore, to further determine whether loss of Pfn1 impacts spontaneously developing breast cancer, we crossed Pfn1flox/flox mice into an MMTV-PyMT:MMTV-iCre background (PyMT: Polyoma virus middle T antigen) to conditionally delete Pfn1 gene in the mammary gland. We chose MMTV-PyMT tumorigenesis model for several reasons. First, PyMT mammary tumors initially develop from a single or few apposed foci in the mammary gland reflecting the initiation of human disease. Second, PyMT tumors progress through 4 distinctly identified stages (hyperplasia, adenoma, early and late stage carcinoma) mimicking the natural progression of human breast cancer. Third, MMTV-PyMT mouse model resembles human luminal breast cancer cells thus allowing us to investigate Pfn1's role in mammary tumorigenesis in a different disease sub-type settingCitation20). PyMT-positive female off-springs of various Pfn1 genotypes (PyMT:Pfn1+/+, PyMT:Pfn+/− and PyMT:Pfn1−/−) were isolated based on PCR analyses of PyMT, Pfn1 (wild-type [WT] allele: 240 bp amplicon; floxed allele: 340 bp amplicon) and iCre on DNA isolated from the tails of the animals (; note that regardless of the status [WT or floxed] of Pfn1 alleles, mice that are Cre-negative are termed as Pfn1+/+ here). All PyMT mice formed palpable tumors which progressed to carcinoma. Kaplan-Meier survival analyses showed that PyMT:Pfn1−/− animals had significantly longer survival (time taken for the largest tumor to reach 2.0 cm in size) than PyMT:Pfn1+/+ (p = 0.02) (). Consistent with these survival data, the total tumor burden at the time of sacrifice in PyMT:Pfn1−/− animals was also 33% lower than that calculated for PyMT:Pfn1+/+ animals, and this difference was fairly close to statistically significant (p = 0.09) (). We found no statistically significant difference in either survival or tumor burden between PyMT:Pfn1+/+ andPyMT:Pfn1+/− animals, suggesting that heterozygosity of Pfn1 does not cause any growth-related phenotype in vivo.
Figure 3. Effect of loss of Pfn1 on tumorigenicity in MDA-231 xenograft and MMTV:PyMT breast cancer models. (A) A table showing primary mammary tumor induction frequency following orthotopic inoculation of sh-control vs sh-Pfn1 MDA-231 cells in nude mice. (B) Top panel: A schematic diagram showing the positioning of lox sites in Pfn1flox/flox mouse with respect to the exons (indicated by E1, E2 and E3) and the PCR primers used for genotyping (primers 1 and 2) and confirmation of allele excision (primers 1 and 3) in PyMT+ mice of various Pfn1 genotypes. bottom panel: Pfn1- and Cre PCRs performed with tail-clip DNA. (C) A Kaplan-Meier curve showing relative survival of PyMT+ mice of various Pfn1 genotypes (survival was measured based on the number of days for largest tumor to reach 2.0 cm in size). (D) A bar graph showing the average tumor burden in PyMT+ mice of various Pfn1 genotypes. (E) Pfn1 PCRs performed with tumor-derived DNA (240 bp: wild-type (WT) allele amplicon, 340 bp: floxed allele amplicon, 700 bp: excision-specific amplicon). Note that all Cre-negative mice were considered to be Pfn1+/+ regardless of the nature of Pfn1 alleles (WT/flox, flox/flox, WT/WT). (F-G) A representative immunoblot depicting (F) and a bar graph summarizing (G) the relative Pfn1 expressions in PyMT tumors of the indicated Pfn1 genotypes. Three random tumors were analyzed per genotype; GAPDH immunoblot served as the loading control (data summarized from a pool of 18 random tumor samples from 6 animals of each genotype). (H) Representative IHC images PyMT mammary tumors of the indicated Pfn1 genotypes (Pfn1: red; DAPI: blue) reveal mosaic Pfn1 staining in Pfn1−/− tumors (the area outlined by a dashed line represents a region of the tumor comprised of Pfn1-null cells; bar -100 μm). Values are presented as mean ± SD (*p < 0.05, ***p < 0.001).
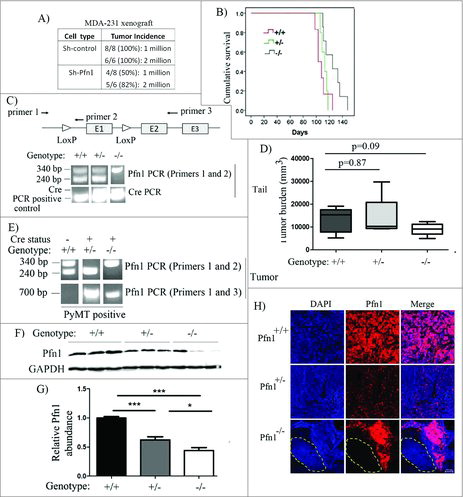
It is known that MMTV-Cre functions incompletely in mammary epithelium (MMTV promoter is active in only 70% of mammary epithelial cells).Citation21,Citation22 In this model, inactivation of tumor-initiating and/or -promoting genes leads to progressive elimination of cells that are null for those genes due to competitive outgrowth of population of cells which remain proficient for those genes (these cells arise due to either escape from Cre expression or incomplete recombination), and as a result, genetic mosaicism occurs.Citation23 In the conventional MMTV-PyMT:MMTV-Cre transgenic mouse model (as used herein), even though PyMT and Cre are driven by MMTV promoter, they are not linked on the same allele allowing cells to express PyMT without expressing Cre or vice-versa. Therefore, in this model, complete inactivation of gene that is essential for tumorigenesis can cause positive selection for PyMT-transformed cells that are negative for Cre expression, as seen previously upon conditional inactivation of several genes.Citation24,Citation25 Our PCR, immunoblot and IHC analyses of PyMT tumors for Pfn1, as elaborated below, collectively suggest that homozygous but not heterozygous deletion of Pfn1 leads to positive selection of Pfn1-proficient cells. First, in the case of PyMT:Pfn+/- tumors, PCR analyses revealed complete loss of 340 bp amplicon and appearance of 700 bp amplicon (this knockout-specific band results only if the floxed allele is excised) suggesting efficient excision of the floxed allele (). As per immunoblot analyses, the average Pfn1 expression of PyMT:Pfn1+/− tumors was found to be 40% lower (p < 0.0001) than PyMT:Pfn1+/+ tumors (). Since heterozygosity should theoretically result in 50% reduction of protein expression, Pfn1 expression downregulation correlated reasonably well with the genotype status in PyMT:Pfn1+/− tumors. Consistent with these data, IHC of PyMT:Pfn1+/− tumors revealed uniform Pfn1 downregulation when compared with PyMT:Pfn1+/− tumors (). Collectively, these data suggest that PyMT:Pfn+/- tumors derived almost exclusively from cells that are heterozygous for Pfn1 gene. By contrast, PCR analyses of PyMT:Pfn−/− tumors revealed genetic mosaicism of Pfn1 as demonstrated by the presence of both 340 bp and 700 bp amplicons (). Accordingly, immunoblots of PyMT:Pfn1−/− tumor extracts consistently showed bands that were indicative of either negligible Pfn1 expression (suggesting presence of Pfn1-null cells in at least these sampled tumor regions) or Pfn1 expression comparable to or higher than the level of PyMT:Pfn1+/- tumors. Although the average Pfn1 expression of PyMT:Pfn1−/− tumors was statistically significant lower than either PyMT:Pfn1+/+ or PyMT:Pfn1+/− tumors, the overall protein reduction clearly did not correlate with the expected inactivation status (). In further support of these findings, IHC data revealed highly mosaic Pfn1 expression status in PyMT:Pfn−/− tumors where the expression ranged from undetectable to a level as high as that seen in PyMT:Pfn+/+ tumors (). Since the overall phenotype of PyMT:Pfn1−/− tumors is likely determined by both Pfn1-null and –proficient cells, and the number of animals was small, a p-value fairly close to but not less than 0.05 signifying the difference in the mean tumor burden between PyMT:Pfn1+/+ and PyMT:Pfn1−/− tumor () was not at all surprising. Prominent selection of Pfn1-proficient cells in Pfn1−/− genetic background leading to mosaic Pfn1 status in PyMT:Pfn1−/− tumors is consistent with a scenario in which Pfn1-null cells are out-competed by Pfn1-proficient cells in terms of their tumor-initiating ability. Although indirect, these data support a general contention that a complete lack of Pfn1 may not be conducive for efficient tumor-initiating capability of breast cancer cells, similar to the effect of elevated level of Pfn1 as reported previously.Citation2,Citation4 Based on these findings, we conclude that a balanced level of Pfn1 promotes efficient tumor-initiation by breast cancer cells.
Discussion
Stemness is a key biologic feature that is associated with greater tumor-initiating potential of cancer cells. In this study, we demonstrated for the first time that Pfn1 is an important determinant of stemness of MDA-231 triple-negative breast cancer cells. We showed that the mammosphere-forming efficiency (a common experimental readout for stem-like characteristics of tumor cells) of MDA-231 cells was reduced when Pfn1 expression was either depleted or elevated, suggesting requirement of a balanced level of Pfn1 for optimal maintenance of stem-like state of MDA-231 cells. These observations seem to be consistent with previously established Pfn1's ability to completely suppress tumor-initiation capability of MDA-231 cells when overexpressedCitation3 and the present finding of reduced tumor-initiating efficiency of xenografted MDA-231 cells upon depletion of Pfn1. Since stem-like attributes enhance metastatic competency of cancer cells, this may also explain why metastatic outgrowth from isolated extravasated MDA-231 cells in the lungs is severely impaired upon depletion of Pfn1 as recently demonstrated by our group.Citation3 As the present study is exclusively focused on Pfn1s effect on stem-like traits of MDA-231 cells (a limitation of this study), we do not know whether Pfn1 impacts the stemness of other triple-negative breast cancer cells in a similar manner — this needs to be investigated in the future.
How genetic loss of Pfn1 impacts cancer initiation has been studied before. Although indirect, the findings from our PyMT-based knockout mouse model studies tend to support the general idea of the requirement of some minimum level of Pfn1 for efficient spontaneous initiation of tumors of luminal subtype from either single or apposed foci in the mammary gland. However, an improved transgenic model capable of triggering coupled expression of PyMT and Cre (eliminates the problem of genetic mosaicism) in an inducible manner (such as one recently developed by the Muller groupCitation21 with additional xenografts studies with luminal breast cancer cells will be needed in the future for a definitive proof of this tenet.
Attenuation of stemness-enriched pool of MDA-231 upon stable knockdown of Pfn1 expression as shown herein is consistent with previously reported reduced in vivo retention of normal HSC upon genetic knockout of Pfn1.Citation7 In the context of normal HSC, Pfn1 depletion compromises stem cell retention via promoting apoptosis and induction of cell-cycle quiescence. Since we do not see any general evidence of increased cell death in MDA-231 cells upon knockdown of Pfn1 (data not shown), we speculate that at least in in vitro culture, increased apoptosis does not account for reduced mammosphere forming efficiency of Pfn1-depleted MDA-231 cells compared to their control counterparts.
Although mammosphere formation was reduced under both depleted and elevated conditions of Pfn1, interestingly, mammosphere growth (an indicator of self-renewal ability of stem-like cells) was affected only when Pfn1 expression was elevated. This was not totally surprising given that overexpression of Pfn1 had a much more robust effect on the expressions of CSC-related genes when compared with that elicited by Pfn1 depletion. Out of the 84 genes we probed, we only found 4 genes that showed opposite trends of gene expression upon overexpression and knockdown of Pfn1 expression, and we performed immunoblot analyses of 2 of these genes (MUC1 and STAT3) to validate Pfn1-dependent changes at the protein level. Of particular importance is the prominent 6-fold reduction and 12-fold increase in MUC1 mRNA abundance upon depletion and overexpression of Pfn1, respectively. It has been shown that MUC1-C, the transmembrane C-terminal domain of MUC1 generated by the natural autocleavage of the full-length protein with oncogenic function, is expressed at an elevated level in the stem-like subpopulation of breast cancer cells. Mammosphere-forming efficiency of breast cancer cells is enhanced upon overexpression of MUC1-C, and conversely, it is reduced upon knockdown of MUC1 suggesting that MUC1 is an important promoter for stemness of breast cancer cells.Citation26 Although we were able to validate the increase of MUC1 expression at the protein level upon Pfn1 overexpression, the fold-increase in the protein level (∼2-fold) was significantly lower than the corresponding fold-change of the mRNA level. One potential explanation for this discrepancy could be that our immunoblot analyses were performed with the whole cell lysate and therefore did not analyze the secreted fraction of MUC1. It is also known that MUC1 undergoes alternative splicing to generate several variants.Citation27 Since the antibody used for immunoblot analyses targets the APDTR epitope in the VNTR (variable number of tandem repeats) region of MUC1 and does not detect those alternatively spliced isoforms devoid of the VNTR region (usually <50 kDa), it is also possible that we were not able to assess these splice variants. Therefore, if Pfn1 knockdown somehow affected either the secreted fraction or any of the alternative spliced forms of MUC1, it would be undetected by the present analysis and this, if true, could potentially explain why we were unable to see changes in MUC1 expression at the protein level from the whole cell lysate despite a dramatic 6-fold decrease in the mRNA level upon knockdown of Pfn1. Although MUC1 promotes self-renewal capacity of breast cancer stem cells,Citation26 lack of growth-related phenotype could be due to the balancing act resulting from upregulation of several self-renewal promoting genes such as integrin β1, STAT3 and Fzd7 (Frizzled – a key mediator of Wnt signaling) in Pfn1 knockdown setting. Interestingly, a recent study reported downregulation of cell-surface integrin β1 expression and Wnt signaling in a bladder cancer cell line upon knockdown of Pfn1 expression.Citation28 While this may suggest apparent contradictory effects of Pfn1 knockdown between breast and bladder cancer cells, because of differences in experimental parameters between the 2 studies (e.g., surface protein expression vs mRNA level of integrin β1) it is difficult to reconcile these results. Cell-specific differences in the results will not be totally surprising given that Pfn1 has context-specific effects in normal vs cancer cells and even between different cancer cell lines.Citation29,Citation30,Citation31
Finally, CSCs offer resistance to therapy and are thought to cause relapse of breast cancer cells. Therefore modulating the stemness of breast cancer cells through perturbation of Pfn1 could be a possible strategy to sensitize breast cancer cells to chemotherapy in vivo. In fact, in cell culture model, there is already evidence for increased susceptibility of breast cancer cells to cytotoxic agents upon overexpression of Pfn1.Citation32,Citation33
Material and methods
Animal experiments, DNA isolation and protein extraction
Orthoptopic implantation assays involving MDA-231 cells have been described previously.Citation3 For conditional knockout of Pfn1, Pfn1flox/flox mice (C57B6xCBA)Citation34 were backcrossed 5 generations into FVB background before breeding into MMTV:PyMT-MMTV:iCre background. Animals were killed when the diameter of the largest tumor reached 2.0 cm. All animal experiments were performed according to the University of Pittsburgh Institutional animal welfare guidelines. Genomic DNA was isolated using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI) according to manufacturer's protocol before subjecting to PCR analyses (see Table S1 for PCR reaction details). Tumor lysate was prepared with RIPA buffer with 0.2% SDS, homogenized 2X for 15 sec and clarified by centrifugation at 18000 g for 30 min before SDS-PAGE/ immunoblot analyses. (see Table S2 for source and concentration of different antibodies). Pfn1/DAPI staining of tumor histosections was performed according to our published protocol.Citation35
Cell culture
Generation and culture of MDA-231 cell lines stably expressing GFP, GFP-Pfn1, control- and Pfn1-shRNAs have been described previously.Citation3,Citation4
Mammosphere assay
MDA-231 cells were trypsinized, filtered through a 40 μm cell strainer, and then plated in 12 well ultra-low attachment plates at a density of 1000 cells/well in serum-free mammary epithelium basal medium (Lonza, Walkersville, MD) supplemented with 1% antibiotics, 2% B27 (Life Technologies, Grand Island, NY), 5 μg/mL Insulin, 1 μg/mL hydrocortisone (Sigma-Aldrich, St. Louis, MO), 20 ng/mL EGF (R&D Systems, Minneapolis, MN), 20 ng/mL bFGF (StemCell Technologies, Inc., Vancouver, BC) and β-mercaptoethanol (1:25,000). After 8 d incubation, the mammosphere number was counted under an inverted microscope with a 10X objective and the mammopsherere size was measured.
Cancer Stem Cell (CSC) gene expression analyses
Total RNA was extracted from MDA- 231 cells using RNeasy Mini Kit (Qiagen, Valencia, CA). The first strand cDNA synthesis was conducted using RTCitation2 First Strand Kit and PCR was performed using in Human Cancer Stem Cells PCR Array (Qiagen) according to the manufacturer's instructions.
Statistical analysis
Statistical differences were assessed by Student's T-test or one way ANOVA followed by Tukey post-hoc test, and a p-value of less than 0.05 was considered significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
CJ, ZJ, MJ, SC and SHK performed experiments, analyzed the data, and wrote the manuscript. RB generated the original Pfn1 floxed mouse and provided intellectual input to the manuscript. JC provided MMTV:PyMT mouse and intellectual input in the study design, SS provided intellectual input in the study design. PR conceived the study, designed experiments, and wrote the manuscript.
KCCY_S_1346759.docx
Download MS Word (154.7 KB)Acknowledgments
We thank Dr. Reinhard Fassler (Max Planck Institute) for the generous gift of Pfn1flox/flox mouse. We also thank Drs. Anda Vlad and Lixin Zhang (University of Pittsburgh) for providing us the MUC1 antibody and helpful suggestions on MUC1 immunoblot.
Additional information
Funding
References
- Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, Simpson KJ, Lim J, Gertler FB, Hartwig JH, et al. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell 2012; 22:615–30; PMID:23153535; https://doi.org/10.1016/j.ccr.2012.09.027
- Janke J, Schluter K, Jandrig B, Theile M, Kölble K, Arnold W, Grinstein E, Schwartz A, Estevéz-Schwarz L, Schlag PM, et al. Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin 1. J Exp Med 2000; 191:1675–86; PMID:10811861; https://doi.org/10.1084/jem.191.10.1675
- Ding Z, Joy M, Bhargava R, Gunsaulus M, Lakshman N, Miron-Mendoza M, Petroll M, Condeelis J, Wells A, Roy P. Profilin-1 downregulation has contrasting effects on early vs late steps of breast cancer metastasis. Oncogene 2014; 33:2065–74; PMID:23686314; https://doi.org/10.1038/onc.2013.166
- Zou L, Jaramillo M, Whaley D, Wells A, Panchapakesa V, Das T, Roy P. Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br J Cancer 2007; 97:1361–71; PMID:17940506; https://doi.org/10.1038/sj.bjc.6604038
- Wittenmayer N, Jandrig B, Rothkegel M, Schlüter K, Arnold W, Haensch W, Scherneck S, Jockusch BM. Tumor suppressor activity of profilin requires a functional actin binding site. Mol Biol Cell 2004; 15:1600–8; PMID:14767055; https://doi.org/10.1091/mbc.E03-12-0873
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014; 14:275–91; PMID:24607403; https://doi.org/10.1016/j.stem.2014.02.006
- Zheng J, Lu Z, Kocabas F, Böttcher RT, Costell M, Kang X, Liu X, Deberardinis RJ, Wang Q, Chen GQ, et al. Profilin 1 is essential for retention and metabolism of mouse hematopoietic stem cells in bone marrow. Blood 2014; 123:992–1001; PMID:24385538; https://doi.org/10.1182/blood-2013-04-498469
- Kim MJ, Lee YS, Han GY, Lee HN, Ahn C, Kim CW. Profilin 2 promotes migration, invasion, and stemness of HT29 human colorectal cancer stem cells. Biosci Biotechnol Biochem 2015; 79:1438–46; PMID:25964982; https://doi.org/10.1080/09168451.2015.1043118
- Zou L, Hazan R, Roy P. Profilin-1 overexpression restores adherens junctions in MDA-MB-231 breast cancer cells in R-cadherin-dependent manner. Cell Motil Cytoskeleton 2009; 66:1048–56; PMID:19593789; https://doi.org/10.1002/cm.20407
- Joy M, Gau D, Castellucci N, Prywes R, Roy P. The Myocardin-related transcription factor MKL co-regulates the cellular levels of two profilin isoforms. J Biol Chem 2017; 292, 11777–11791; PMID:28546428; https://doi.org/10.1074/jbc.M117.781104
- Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R Jr, Badve S, Nakshatri H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res: BCR 2006; 8:R59; PMID:17062128; https://doi.org/10.1186/bcr1610
- Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci U S A 2010; 107:21737–42; PMID:21098263; https://doi.org/10.1073/pnas.1007863107
- Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R, Martin AG. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012; 31:1354–65; PMID:21822303; https://doi.org/10.1038/onc.2011.338
- Taddei I, Deugnier MA, Faraldo MM, Petit V, Bouvard D, Medina D, Fässler R, Thiery JP, Glukhova MA. Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nature Cell Biol 2008; 10:716–22; PMID:18469806; https://doi.org/10.1038/ncb1734
- Chen L, Fan J, Chen H, Meng Z, Chen Z, Wang P, Liu L. The IL-8/CXCR1 axis is associated with cancer stem cell-like properties and correlates with clinical prognosis in human pancreatic cancer cases. Sci Rep 2014; 4:5911; PMID:25081383; https://doi.org/10.1038/srep05911
- Chung SS, Aroh C, Vadgama JV. Constitutive activation of STAT3 signaling regulates hTERT and promotes stem cell-like traits in human breast cancer cells. PloS One 2013; 8:e83971; PMID:24386318; https://doi.org/10.1371/journal.pone.0083971
- Lu H, Clauser KR, Tam WL, Fröse J, Ye X, Eaton EN, Reinhardt F, Donnenberg VS, Bhargava R, Carr SA, et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol 2014; 16:1105–17; PMID:25266422; https://doi.org/10.1038/ncb3041
- Alam M, Rajabi H, Ahmad R, Jin C, Kufe D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget 2014; 5:2622–34; PMID:24770886; https://doi.org/10.18632/oncotarget.1848
- Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med 2014;20:332–42; PMID:24667139; https://doi.org/10.1016/j.molmed.2014.02.007
- Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 2003; 163:2113–26; PMID:14578209; https://doi.org/10.1016/S0002-9440(10)63568-7
- Rao T, Ranger JJ, Smith HW, Lam SH, Chodosh L, Muller WJ. Inducible and coupled expression of the polyomavirus middle T antigen and Cre recombinase in transgenic mice: an in vivo model for synthetic viability in mammary tumour progression. Breast Cancer Res: BCR 2014; 16:R11; PMID:24457046; https://doi.org/10.1186/bcr3603
- Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res 2001; 10:545–53; PMID:11817542; https://doi.org/10.1023/A:1013063514007
- Lu P, Ewald AJ, Martin GR, Werb Z. Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev Biol 2008; 321:77–87; PMID:18585375; https://doi.org/10.1016/j.ydbio.2008.06.005
- White DE, Kurpios NA, Zuo D, Hassell JA, Blaess S, Mueller U, Muller WJ. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell 2004; 6:159–70; PMID:15324699; https://doi.org/10.1016/j.ccr.2004.06.025
- Lahlou H, Sanguin-Gendreau V, Zuo D, Cardiff RD, McLean GW, Frame MC, Muller WJ. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci U S A 2007; 104:20302–7; PMID:18056629; https://doi.org/10.1073/pnas.0710091104
- Alam M, Ahmad R, Rajabi H, Kharbanda A, Kufe D. MUC1-C oncoprotein activates ERK–>C/EBPbeta signaling and induction of aldehyde dehydrogenase 1A1 in breast cancer cells. J Biol Chem 2013; 288:30892–903; PMID:24043631; https://doi.org/10.1074/jbc.M113.477158
- Zhang L, Vlad A, Milcarek C, Finn OJ. Human mucin MUC1 RNA undergoes different types of alternative splicing resulting in multiple isoforms. Cancer Immunol Immunother 2013; 62:423–35; PMID:22941036; https://doi.org/10.1007/s00262-012-1325-2
- Frantzi M, Klimou Z, Makridakis M, Zoidakis J, Latosinska A, Borràs DM, Janssen B, Giannopoulou I, Lygirou V, Lazaris AC, et al. Silencing of Profilin-1 suppresses cell adhesion and tumor growth via predicted alterations in integrin and Ca2+ signaling in T24M-based bladder cancer models. Oncotarget 2016; 7:70750–68; PMID:27683119
- Cheng YJ, Zhu ZX, Zhou JS, Hu ZQ, Zhang JP, Cai QP, Wang LH. Silencing profilin-1 inhibits gastric cancer progression via integrin beta1/focal adhesion kinase pathway modulation. World J Gastroenterol 2015; 21:2323–35; PMID:25741138; https://doi.org/10.3748/wjg.v21.i8.2323
- Shen K, Xi Z, Xie J, Wang H, Xie C, Lee CS, Fahey P, Dong Q, Xu H. Guttiferone K suppresses cell motility and metastasis of hepatocellular carcinoma by restoring aberrantly reduced profilin 1. Oncotarget 2016; 7:56650–63; PMID:27494863
- Ding Z, Bae YH, Roy P. Molecular insights on context-specific role of profilin-1 in cell migration. Cell Adh Migr 2012; 6:442–9; https://doi.org/10.4161/cam.21832
- Zou L, Ding Z, Roy P. Profilin-1 overexpression inhibits proliferation of MDA-MB-231 breast cancer cells partly through p27kip1 upregulation. J Cell Physiol 2010; 223:623–9; PMID:20143334.
- Yao W, Cai X, Liu C, Qin Y, Cheng H, Ji S, Xu W, Wu C, Chen T, Xu J, et al. Profilin 1 potentiates apoptosis induced by staurosporine in cancer cells. Curr Mol Med 2013; 13:417–28; PMID:23331014
- Bottcher RT, Wiesner S, Braun A, Wimmer R, Berna A, Elad N, Medalia O, Pfeifer A, Aszódi A, Costell M, et al. Profilin 1 is required for abscission during late cytokinesis of chondrocytes. The EMBO J 2009; 28:1157–69; PMID:19262563; https://doi.org/10.1038/emboj.2009.58
- Gau DM, Lesnock JL, Hood BL, Bhargava R, Sun M, Darcy K, Luthra S, Chandran U, Conrads TP, Edwards RP, et al. BRCA1 deficiency in ovarian cancer is associated with alteration in expression of several key regulators of cell motility - A proteomics study. Cell Cycle 2015; 14:1884–92; PMID:25927284; https://doi.org/10.1080/15384101.2015.1036203
