Microtubules are organized by 2 peripheral centrosomes and one centrally located plate of chromosomes to create the mitotic spindle. The function of this spindle is to faithfully segregate pairs of chromosomes into 2 daughter cells. This bipolar spindle is a universal feature of animal cells with one primary exception: cancer cells, which frequently evolve excess centrosomes in many types of human cancers.Citation1 The selective advantages of centrosome amplification to cancer cells are thought to involve increasing rates of chromosomal alterations (the mutator phenotype hypothesis) and enhancing cancer cell invasiveness through alterations in cell polarity.Citation2
Despite these advantages of centrosome amplification to cancer cells, excess centrosomes complicate mitosis since the cell division machinery is adapted to manage only 2 centrosomes and form a bipolar spindle. Research has demonstrated that cancer cells use a novel mechanism to divide with excess centrosomes: they cluster their centrosomes together into 2 foci to generate a pseudo-bipolar spindle structure.Citation1 Since this centrosome clustering process only occurs in cancer cells, this could represent an “Achilles’ Heel” for highly specifically targeting cancer cells while sparing normal cells.
Previous work has elucidated some of the factors involved in clustering centrosomes.Citation1,3,4 For instance, Kwon et al.Citation3 performed an RNAi screen in Drosophila cells and found that cell adhesion, actin and molecular motors are all important for centrosome clustering. In addition, several chemical compounds have been identified as inhibitors of centrosome clustering. Kawamura et al.Citation4 performed a high content screen of compound libraries for centrosome clustering inhibitors and identified multiple promising drug candidates. This demonstrates that cancer cells can be specifically targeted using centrosome clustering inhibitors.
In our recent study,Citation5 a compound library screen for inhibitors of centrosome clustering lead us to Stat3, an oncogene that is primarily involved in transcriptional regulation of apoptosis, inflammation and invasiveness. We uncovered a non-transcriptional role for Stat3 in centrosome clustering and focused on determining how Stat3 could regulate centrosome clustering.
Previous work showed that Stat3 interacts with and inhibits the microtubule depolymerase Stathmin.Citation6 Since microtubules are involved in centrosome clustering, Stathmin seemed like a plausible candidate in our pathway. Our data showed that Stathmin interacts with Stat3 and is downstream of Stat3 but curiously, bulk changes in microtubule depolymerisation do not affect the Stat3 centrosome clustering pathway. This suggests that Stathmin is primarily acting independent of its microtubule depolymerase function. To explore what could be downstream of Stathmin, we next screened a kinase inhibitor library for inhibitors of centrosome clustering and cross-checked whether Stat3 inhibitors blocked activation of these candidate kinases. We discovered that the mitosis-specific central regulator of centrosome maturation, PLK1, is both required for centrosome clustering and is regulated by our Stat3-Stathmin pathway. This suggests that PLK1 is downstream of Stat3 and Stathmin.
PLK1 has been previously implicated in centrosome clustering in endothelial cells, where it induces clustering by increasing γ-Tubulin levels.Citation7 γ-Tubulin nucleates microtubules at centrosomes and helps determine the number of microtubules per centrosome. Previous research demonstrated that total γ-Tubulin levels do not increase when centrosomes are amplified which means that each extra centrosome has less γ-Tubulin available to associate with. The model that these researchers proposed is that reduced γ-Tubulin levels per centrosome cause a decrease in microtubules per centrosome and since microtubules position centrosomes in the cell, cells with amplified centrosomes have reduced ability to correctly position and cluster centrosomes. We observed that Stat3 inhibition reduced γ-Tubulin levels at the centrosome as well as reduced the number of mitotic centrosomes with long astral microtubules. Our current model is that Stat3 activates PLK1 to increase γ-Tubulin levels, restoring microtubule-dependent centrosome positioning in cells with centrosome amplification and allowing centrosome clustering to occur. This suggests that part of the function of Stat3 overexpression in cancer could be to induce centrosome clustering ().
Figure 1. Stat3-dependent centrosome clustering pathway and a proposed model of the sequence of events that leads to centrosome clustering in cancer cells.
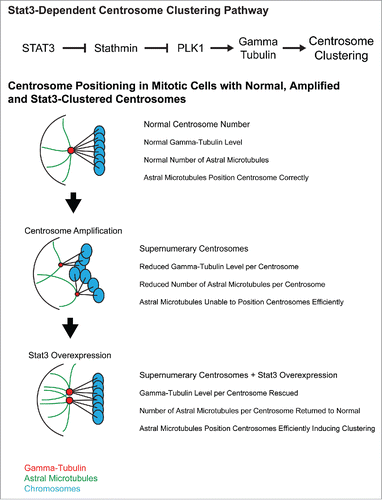
This study is the first demonstration of a function for Stat3 in mitosis and it is currently unclear how this pathway is regulated upstream of Stat3. Stat3 function is largely regulated by nuclear-cytoplasmic shuttling and since the nucleus breaks down during cell division and shuttling does not occur, the upstream regulation of Stat3 during mitosis could be significantly different from canonical Stat3 activation.
There are currently multiple clinical trials of Stat3 inhibitors. In our recent paper, we demonstrated that Stat3 inhibitors are significantly more effective in inhibiting cell growth in cancer cells with centrosome amplification relative to cancer cells with normal centrosome number. We are planning to score centrosome amplification in human tumor samples to determine whether patients with tumors that have amplified centrosomes have better Stat3 inhibitor treatment outcomes. If Stat3 inhibitors do block centrosome clustering in patients, this would represent the first time a centrosome clustering inhibitor has been tested in patients, creating an important milestone in the development of a centrosome clustering inhibitor for use in targeting cancer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science 2005; 307:127-9; PMID:15637283; https://doi.org/10.1126/science.1104905
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Théry M, Pellman D. Oncogene-like induction of cellular invasion from centrosome amplification. Nature 2014; 510:167-71; PMID:24739973; https://doi.org/10.1038/nature13277
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 2008; 22:2189-203; PMID:18662975; https://doi.org/10.1101/gad.1700908
- Kawamura E, Fielding AB, Kannan N, Balgi A, Eaves CJ, Roberge M, Dedhar S. Identification of novel small molecule inhibitors of centrosome clustering in cancer cells. Oncotarget 2013; 4:1763-76; PMID:24091544; https://doi.org/10.18632/oncotarget.1198
- Morris EJ, Kawamura E, Gillespie JA, Balgi A, Kannan N, Muller WJ, Roberge M, Dedhar S. Stat3 regulates centrosome clustering in cancer cells via Stathmin/PLK1. Nat Commun 2017; 8:15289; PMID:28474672; https://doi.org/10.1038/ncomms15289
- Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, Cao X. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol 2006; 172:245-57; PMID:16401721; https://doi.org/10.1083/jcb.200503021
- Kushner EJ, Ferro LS, Liu JY, Durrant JR, Rogers SL, Dudley AC, Bautch VL. Excess centrosomes disrupt endothelial cell migration via centrosome scattering. J Cell Biol 2014; 206:257-72; PMID:25049273; https://doi.org/10.1083/jcb.201311013
