Epigenetic modifications organize the genome into open (“euchromatin”) and closed (“heterochromatin”) chromatin domains, which drive translation of genetic information into specific outputs in different cell types. The post-translational modifications of histones have emerged as a critical component of epigenetic marking systems. Histone modifications not only dictate the organization of chromatin into distinct domains, but also transmit epigenetic memory during cell division. However, DNA replication poses a challenge for preserving epigenetic memory, requiring chromatin associated histone modifications to be restored following chromatin disruption.
Histone modifications, such as histone 3 lysine 9 methylation (H3K9me) that marks heterochromatin, have been widely studied including in the fission yeast Schizosaccharomyces pombe. The H3K9 methyltransferase Clr4/Suv39h is recruited by DNA sequence- or RNA-dependent “de novo” nucleation mechanisms to establish heterochromatin. Once established, heterochromatin itself provides additional epigenetic specificity (“epigenetic-template”) that can propagate heterochromatic structures in a self-templating manner in cis,Citation1 a role more evident when de novo nucleation mechanisms are impaired. We have shown that the ability of Clr4/Suv39h to methylate H3K9 (“write”) and also bind to H3K9me (“read)”) establishes an elegant feedback “loop” critical for epigenetic inheritance of heterochromatin.Citation2 Importantly, parental histones are carriers of epigenetic memory and must be faithfully segregated upon cell division, yet the mechanisms have remained unclear.
Indeed, few components involved in the inheritance of epigenetic states have been uncovered. Using a specially designed genetic screen, we recently identified the SNF2 family ATP-dependent chromatin remodeler, Fft3 (Fission Fun Thirty), as required for heterochromatin maintenance in dividing cells, but dispensable for de novo assembly.Citation3 Fft3 is a homolog of SMARCAD1 (human) and FUN30 (budding yeast) implicated in heterochromatin assembly Citation4 (and references therein). We found that Fft3 suppresses histone turnover and ensures faithful transmission of parental histones to maintain heterochromatin in daughter cells. Fft3 also precludes histone turnover at difficult-to-replicate euchromatic loci to prevent formation of aberrant structures and to allow passage of replication forks.
De novo assembly mechanisms acting concomitantly with epigenetic-templated maintenance hinder the identification of inheritance factors. We performed a screen using a yeast strain defective in de novo assembly of heterochromatin at the mating-type (mat) locus. Disruption of the major nucleation mechanism at mat creates a metastable heterochromatic locus. The epigenetic ON (expressed) and OFF (silenced) states, which differ in H3K9me levels in otherwise genetically identical cells, are faithfully inherited in cis upon cell division, even when combined in the same nuclear environment of diploid cells.Citation1 The conserved heterochromatin protein Swi6/HP1 maintains the silenced state, and its transient overexpression can heritably convert the ON epigenetic state to OFF.Citation5 Interestingly, Swi6/HP1 mediated ON to OFF switching requires Fft3 (our unpublished data). Indeed, we found that Fft3 localizes to all heterochromatic regions and interacts with Swi6.
Fft3 and its mammalian homolog SMARCAD1 associate with replication machinery.Citation3,4 Our evidence suggests that Fft3 suppresses nucleosome turnover and facilitates the transmission of old parental histones ().Citation3 Unlike cells lacking other canonical heterochromatin factors, such as Snf2/Histone deacetylase-containing Repressor Complex (SHREC) that includes the histone deacetylase Clr3, precludes turnover of nucleosomes at heterochromatin during the major G2 phase of the S. pombe cell cycle, Fft3 ensures epigenetic transmission of parental histones in dividing cells. Indeed, cells lacking Fft3 gradually lose heterochromatin with each cell division, indicating that the fidelity of heterochromatin inheritance is compromised. The cumulative increase in histone turnover upon loss of Fft3 and Clr3 suggests overlapping yet distinct roles at heterochromatic loci. Indeed, the peri-centromeric region active for de novo assembly and Clr3 recruitment maintains silencing in the absence of Fft3.
Figure 1. Model depicting inheritance of heterochromatin and genome integrity by chromatin remodeler, Fft3. Fft3 promotes stable maintenance of epigenetic memory by precluding histone turnover at heterochromatic regions and might act together with FACT. Fft3 also suppresses histone turnover and facilitates replication fork progression through specific euchromatic sites. In absence of Fft3, these regions accumulate R-loops, which causes genome instability.
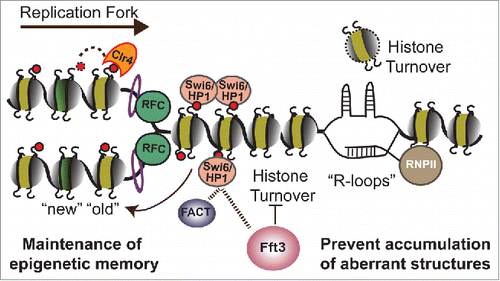
Fft3 also localizes to highly transcribed genes and genes containing internal repeats.Citation3 Moreover, Fft3 was recently reported to interact with transcription machinery;Citation6 whether its localization to genic regions is transcription-dependent is unclear. Remarkably, Fft3 facilitates replication fork progression through these sites, and suppresses nucleosome turnover to prevent RNA-DNA hybrid accumulation ().Citation3 Cells lacking Fft3 experience replication stress, DNA damage and genotoxin sensitivity. Clr4/Suv39h is also required for replication progression through the same loci and for suppressing replication defects and genome instability. Intriguingly, Fft3 along with repressive marks might ensure that these fragile regions are wrapped around histones after the DNA polymerase passes, preventing exposure of DNA to invading RNA and formation of aberrant structures.
Replication and transcription require unwrapping and rebuilding nucleosomes. Chromatin remodelers and histone chaperones are needed to manipulate DNA–histone contacts. Like Fft3, the histone chaperone FACT suppresses nucleosome turnover at highly transcribed genes and aids in redepositing displaced nucleosomes behind the RNA polymerase. FACT also contributes to replication-coupled nucleosome assembly, like Asf1 and CAF-1. Similar to Fft3, FACT also facilitates heterochromatin maintenanceCitation7 and interacts with Swi6. Fft3 may work in conjunction with histone chaperones such as FACT to preserve epigenetic memory by displacing and redepositing nucleosomes in the wake of DNA/RNA polymerases (). A new paradigm also emerges in which suppression of nucleosome turnover prevents formation of structural barriers to promote proper replication and protect genome integrity.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SIS. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232-7. doi:10.1126/science.1076466. PMID:12215653.
- Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381-8. doi:10.1038/nsmb.1406. PMID:18345014.
- Taneja N, Zofall M, Balachandran V, Thillainadesan G, Sugiyama T, Wheeler D, Zhou M, Grewal SIS. SNF2 family protein Fft3 suppresses nucleosome turnover to promote epigenetic inheritance and proper replication. Mol Cell. 2017;66:50-62 e6. doi:10.1016/j.molcel.2017.02.006. PMID:28318821.
- Rowbotham SP, Barki L, Neves-Costa A, Santos F, Dean W, Hawkes N, Choudhary P, Will ER, Webster J, Oxley D, Green CM, Weisz PV, Mermoud JE. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell. 2011;42:285-96. doi:10.1016/j.molcel.2011.02.036. PMID:21549307.
- Nakayama J, Klar AJS, Grewal SIS. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307-17. doi:10.1016/S0092-8674(00)80840-5. PMID:10847685.
- Lee J, Choi ES, Seo HD, Kang K, Gilmore JM, Florens L, Washburn MP, Choe J, Workman JL, Lee D. Chromatin remodeller Fun30Fft3 induces nucleosome disassembly to facilitate RNA polymerase II elongation. Nat Commun. 2017;8:14527. doi:10.1038/ncomms14527. PMID:28218250.
- Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, Allshire RC, Ladurner AG. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol. 2007;17:1219-24. doi:10.1016/j.cub.2007.06.028. PMID:17614284.
