ABSTRACT
microRNAs (miRNAs) play a crucial role in mediation of the cellular sensitivity to ionizing radiation (IR). Previous studies revealed that miR-300 was involved in the cellular response to IR or chemotherapy drug. However, whether miR-300 could regulate the DNA damage responses induced by extrinsic genotoxic stress in human lung cancer and the underlying mechanism remain unknown. In this study, the expression of miR-300 was examined in lung cancer cells treated with IR, and the effects of miR-300 on DNA damage repair, cell cycle arrest, apoptosis and senescence induced by IR were investigated. It was found that IR induced upregulation of endogenous miR-300, and ectopic expression of miR-300 by transfected with miR-300 mimics not only greatly enhanced the cellular DNA damage repair ability but also substantially abrogated the G2 cell cycle arrest and apoptosis induced by IR. Bioinformatic analysis predicted that p53 and apaf1 were potential targets of miR-300, and the luciferase reporter assay showed that miR-300 significantly suppressed the luciferase activity through binding to the 3′-UTR of p53 or apaf1 mRNA. In addition, overexpression of miR-300 significantly reduced p53/apaf1 and/or IR-induced p53/apaf1 protein expression levels. Flow cytomertry analysis and colony formation assay showed that miR-300 desensitized lung cancer cells to IR by suppressing p53-dependent G2 cell cycle arrest, apoptosis and senescence. These data demonstrate that miR-300 regulates the cellular sensitivity to IR through targeting p53 and apaf1 in lung cancer cells.
Introduction
Lung cancer is the leading cause of cancer death in China and non-small cell lung cancer accounts for more than 80% of lung cancer.Citation1,2 Radiotherapy can decrease the lung cancer thoracic recurrence significantly.Citation3 Recently, emerging evidence has highlighted the importance of microRNAs in mediation of the cellular sensitivity to ionizing radiation (IR) and chemotherapy drugs.Citation4-7
microRNAs (miRNAs) are an abundant class of small non-coding RNAs that play important roles in biological processes related to the initiation and progression of various diseases, in particular cancer.Citation8 It has been known that miRNAs function as regulator of oncogenes and tumor suppressor genes.Citation9 Notably, experimental evidence demonstrates that miRNAs affect tumor cell radiosensitivity via influencing DNA damage repair processes and cell death.Citation10,11 For example, miR-18a impairs DNA damage repair and accelerates apoptosis through targeting ATM in colorectal cancer and breast cancer.Citation12,13 miR-185 enhances IR-induced apoptosis and proliferation inhibition via repressing ATR pathway.Citation14 These reports suggest that miRNAs are good potential targets for sensitizing tumor cells to IR. Our previous study showed that miR-300 was involved in the cellular response to IR in gastric cancer.Citation15 Furthermore, Kumar et al revealed that miR-300 expression was upregulated in cisplatin resistant ovarian cells.Citation16 More importantly, both reports indicated that miR-300 may affect p53 signaling pathway to respond to genotoxic stresses and subsequently mediate cell death. However, the underlying mechanism remains largely unexplored.
p53 is a potent tumor suppressor that can inhibit cell growth and induce cell death.Citation17 However, its function is commonly lost in human tumors.Citation18 It is well known that p53 and p53-apaf1 pathways function as central mediators of the cellular DNA damage response incurred by IR or chemotherapy drugs through regulation of DNA damage repair, cell cycle arrest, apoptosis and senescence.Citation19,20 In recent years, miRNAs, such as miR-125bCitation21 and miR-375,Citation22 have been demonstrated to target p53, leading to decreased sensitivity to IR and chemotherapy drugs through rescuing the stress-induced cell cycle arrest and apoptosis. However, the effect of miRNA-p53-apaf1 network on the chemo- and radio-sensitivity remains less well understood.
Here, we investigate the effects of miR-300 on the cellular response to IR in p53 wild type and p53 mutant human lung cancer cells and demonstrate that miR-300-mediated regulation of p53 and apfa1 is crucial for modulating the cellular sensitivity to IR in lung cancer cells.
Results
IR activates miR-300 expression in lung cancer cells
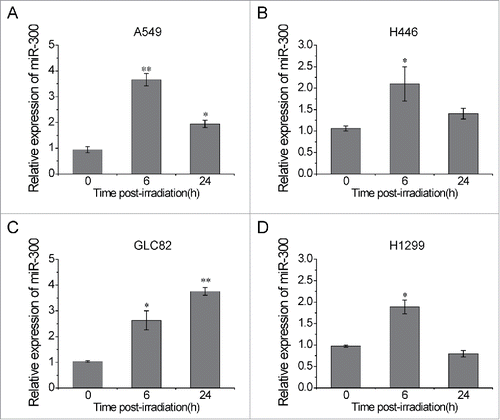
In previous study, decreased expression of miR-300 had been observed in human gastric cancer treated with IR.Citation15 To evaluate the effect of IR on miR-300 expression in lung cancer, human lung cancer cell lines A549, H446, GLC82 and H1299 were exposed to 2 Gy of X-rays. The levels of miR-300 were examined at 0, 6 and 24 h post-irradiation by using qRT-PCR. The results showed that miR-300 levels were significantly increased (approximately 2–4 folds) at 6 h following IR and then slightly declined at 24 h, except for GLC82 cells (), in which miR-300 levels increased continuously after irradiation (). These data indicate that miR-300 involves in the cellular response to IR in lung cancer cells.
Figure 1. IR up-regulates miR-300 expression in lung cancer cells. (A-D) Human lung cancer cells A549 (A), H446 (B), GLC82 (C) and H1299 (D) were exposed to 2 Gy of X-rays. The relative expression levels of miR-300 were evaluated by qRT-PCR at 0, 6, 24 h post-irradiation. * P < 0.05, ** P < 0.01, compared to control.
miR-300 regulates the DNA damage responses to IR
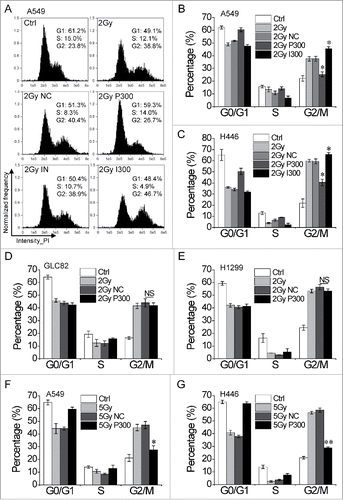
To investigate the role of miR-300 in regulation of the cellular response to IR in lung cancer cells, the intracellular miR-300 levels were up/downregulated by transient transfection with miR-300 mimics (P300) or inhibitor (I300). Cell cycle distribution assay showed that all these four cells were obviously blocked in G2 phase 15 h after irradiated to 2Gy of X-rays. Interestingly, the proportion of cells with G2 phase blockage was dramatically decreased in A549 and H446 cells with miR-300 overexpression (P300) compared to negative control (NC) group (), which was not observed in GLC82 and H1299 cells ( and ). Notably, the G2 cell cycle arrest elicited by IR was aggravated when miR-300 expression was depleted by transfected with I300 in A549 and H446 cells ( and ). Moreover, similar results were obtained 24 h after treated with 5 Gy of X-rays in A549 and H446 cells ( and ).
Figure 2. miR-300 restores IR-induced G2 cell cycle arrest. (A) A549 cells transfected with miR-300 mimics or inhibitor were treated with 2 Gy of X-rays, the cell cycle distribution was measured by flow cytometry. Representative images of each group. (B) The quantification of the data in (A). (C-E) Cell cycle distribution of H446 (C), GLC82 (D) and H1299 (E) cells treated with 2 Gy of X-rays at 15 h post-irradiation. (F-G) Cell cycle distribution of A549 (F) and H446 (G) cells treated with 5 Gy of X-rays at 24 h post-irradiation. Ctrl, control; NC, pre-miRNA negative control; P300, pre-miR-300; IN, miRNA inhibitor negative control; I300, miR-300 inhibitor. * P < 0.05, ** P < 0.01, compared to control; NS, not significant.
Cell cycle arrest provides sufficient time for DNA damage repair.Citation23 53BP1 foci, a widely used marker to detect the DSBs,Citation24–26 were measured at 1 and 12 h post-irradiation. As presented in Fig. S1A and S1B, the 53BP1 foci number increased from ∼2 per cell (sham-irradiated cells) to ∼30 per cell 1 h post-irradiation (cells treated with 0.5 Gy of X-rays irradiation) in A549 cells, but no difference was observed between the irradiated groups. As time went on, the DSBs were gradually cleared in all irradiated groups. However, the 53BP1 foci number in miR-300 overexpression group was greatly less than that in NC group at 12 h post-irradiation, indicating miR-300 elevates the DSBs clearance rate in A549 cells. In addition, the effect of miR-300 on apoptosis induced by IR was tested. It was found that the proportion of apoptotic cells decreased from ∼12% in NC group to ∼8% in P300 group in A549 cells treated with 2 Gy of X-rays (Fig. S1C). Taken together, these data suggest that miR-300 functions as a regulator of the cellular response pathway and its target gene can be activated directly or indirectly by IR.
miR-300 targets p53 and apaf1
The above results have clearly shown that miR-300 diminished IR-induced G2 phase block in A549 and H446 cells not in GLC82 and H1299 cells, but the molecular mechanisms remain unclear. Bioinformatic analysis using microRNA.org, TargetScan and miRDB displayed that p53 and apaf1, which play central roles in cell cycle regulation and apoptosis,Citation27,28 were potential targets of miR-300. One putative miR-300 binding site in p53 mRNA 3′-UTR and three in apaf1 mRNA 3′-UTR were predicted ( and ). As A549 and H446 cells are wild type p53-containing cell lines while p53 in GLC82 or H1299 cells is mutant,Citation29–31 we speculated that miR-300 targets both p53 and apaf1 in p53 wild type cells while in p53 mutant cells miR-300 directly regulates apaf1 expression.
Figure 3. miR-300 targets p53 and apaf1 by binding to mRNA 3′-UTR. (A-B) The sequences of miR-300 and its putative binding sits (rectangle indicated by arrows ) in p53 (A) or apaf1 (B) 3′-UTR. The wild type sequence (WT-P53/APAF1-3′-UTR) or a mutated seed sequence of miR-300-binding site (Mut-P53/APAF1-3′-UTR) were constructed into the luciferase reporter respectively. (C-D) Luciferase reporter containing P53-3′-UTR (C) or APAF1-3′-UTR (D) and miR-300 mimics were co-transfected into A549 cells and the luciferase activity was measured 24 h after transfection. Renilla luciferase activity was used to normalize the firefly luciferase activity. (E) Over-expression of miR-300 down-regulates p53 and apaf1 expression in A549 cells. The levels of p53, p21 and apaf1 were analyzed by western blots 12 h after transfection. (F-H) Over-expression of miR-300 reduces IR-induced p53 and apaf1 expression in A549 (F), H446 (G), H1299 and GLC82 (H) cells. The protein expression levels were measured by western blot 12h after treated with 2 Gy of X-rays. IR, 2 Gy of X-rays irradiation; NC, pre-miRNA negative control; P300, pre-miR-300; +, positive; -, negative. * P < 0.05, compared to NC.
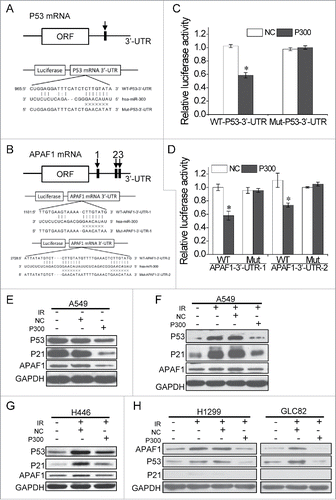
To examine whether miR-300 could bind to the 3′-UTR of p53 or apaf1 mRNA, the wild type and mutant of p53/apaf1 3′-UTR fragments with substitution in the seed region were constructed into the pmirGLO luciferase report system respectively ( and ). Co-transfection of luciferase reporter containing wild type p53 3′-UTR and miR-300 into A549 cells significantly repressed the luciferase activity by approximately 45% (P = 0.012), while suppression of luciferase activity was abolished when a mismatch mutation was introduced in the putative binding sites of p53 3′-UTR (). The same results were obtained using two of apaf1 3′-UTR reporters ().
Next, we validated the inhibition of p53 and apaf1 protein expression by miR-300. As shown in , the expression levels of p53 and apaf1 protein were significantly decreased 12 h after transfection with miR-300 in A549 cells. We further identified the effects of miR-300 on IR-induced p53 or apaf1 expression. The results showed that overexpression of miR-300 specifically suppressed the expression of p53 protein levels at 12 or 24 h post-irradiation ( and S2A). Likewise, ectopic expression of miR-300 suppressed IR-induced p53 and apaf1 upregulation in H446 cells (). Meanwhile, miR-300 overexpression decreased p21 levels, a major transcriptional target of p53 activity,Citation32 in both A549 and H446 cells (), which also indicates p53 gene in A549 or H446 cells can encode a functional protein.
In GLC82 and H1299 cells treated with IR, although p53 expression was detectable by western blot, p21 expression was not activated (), indicating p53 gene is mutant. As we had hypothesized, IR-induced apaf1 expression was also reversed by overexpression of miR-300 in both GLC82 and H1299 cells (). Recent evidence showed that apaf1 is also a transcriptional target of p53 in DNA damage-induced apoptosis,Citation33,34 and our qRT-PCR results showed that overexpression of miR-300 repressed the mRNA expression levels of p21 and apaf1 (Fig. S2B) in A549 cells treated with IR, so apaf1 may be both a direct and indirect target of miR-300 in p53 wild type cells. All data suggest that miR-300 negatively regulates p53 and/or apaf1 in response to IR in lung cancer cells.
miR-300 attenuates the cellular radiosensitivity in lung cancer cells
As p53 and apaf1 play important roles in IR-induce cell cycle arrest and apoptosis,Citation27,28 we checked whether miR-300 influences the cellular sensitivity to IR through interfering p53 or apaf1 expression. siRNAs targeting p53 or apaf1 were introduced to suppress IR-induced p53 and apaf1 activations. p21 and apaf1 protein levels were reduced when p53 was knocked down, while apaf1 depletion did not influence p53 and p21 expression ( and ), indicating apaf1 is a downstream target of p53. Like miR-300 overexpression, p53 knockdown restored IR-induced G2 cell cycle arrest in A549 and H446 cells irradiated to IR ( and ). However, apaf1 knockdown just compromised IR-induced cell cycle arrest in A549 cells but not in H1299 cells (Fig. S3A and S3B), suggesting apaf1-mediated cell cycle arrest may be correlated with p53.
Figure 4. miR-300 modulates radiosensitivity by decreasing p53 and apaf1. (A-B) siRNA targeting p53 (A) or apaf1 (B) interfere its protein expression levels activated by IR. (C-D) miR-300 overexpression or p53 knockdown rescues G2 cell cycle arrest elicited by IR in A549 or H446 cells. Cells transfected with miRNA mimics (NC or P300, 30 nM) or siRNAs (si-NC, or si-p53, 50 nM) were treated with 2 Gy of X-rays The cell cycle distribution was detected 15 h after irradiation by flow cytometry. Representative images of each group in A549 cells (C) and the quantification of each group in H446 cells (D). (E) Survival curves of A549 cells treated with IR after transfected with miR-300 mimics (P300) or p53 siRNA (si-p53). The estimated survival fraction was obtained by fitting to the one-hit multitarget formula at 0, 1, 3 and 5 Gy of X-rays. Data are shown as mean ± SD, the experiment was conducted three times independently. (F) Representative images of colonies in H446 cells treated with 0, 1, 3 and 5 Gy of X-rays. (G) siRNA targeting apaf1 interfere its protein expression levels induced by 5 Gy of X-rays irradiation. The protein levels were analyzed by western blots 12 h after irradiation. (H) The Annexin V-FITC/PtdIns double staining assay in H1299 cells transfected with P300 or si-p53, 24 h following 5 Gy of X-rays irradiation. R, region; PtdIns, propidium iodide; FITC, fluorescein isothiocyanate; IR, 2 Gy of X-rays irradiation; +, positive; -, negative; Ctrl, control; NC, pre-miRNA negative control; P300, pre-miR-300; si-NC, siRNA negative control; si-p53, p53 siRNA; si-apaf1, apaf1 siRNA. * P < 0.05, compared to 2 Gy.
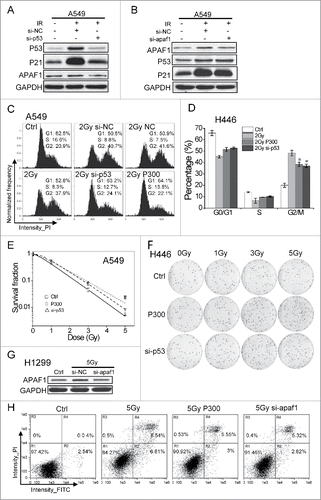
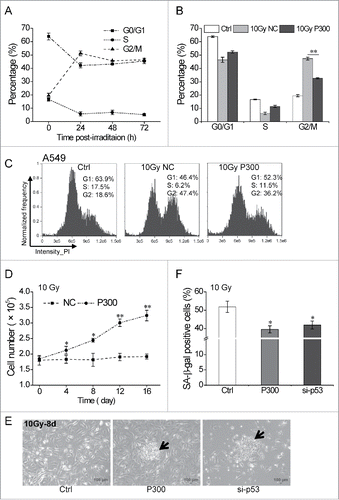
Figure 5. miR-300 reduces p53-dependent senescence induced by IR. (A) High dose irradiation actives cell cycle suspension in A549 cells. A549 cells were exposed to 10 Gy of X-rays, the cell cycle phase was analyzed at 24, 48 and 72 h post-irradiation. (B) miR-300 abolishes cell cycle suspension. The cell cycle phase of A549 cells transfected with miRNA mimics was analyzed at 48 h after irradiation. (C) Representative cell cycle distribution of cells in (B), the data were analyzed with IDEAS Application v6.0. (D) Growth curves of A549 cells treated with 10 Gy of X-rays. Cells were transfected with P300 or NC before irradiation. Graphs represent mean of a triplicate experiment, error bars represent SD. (E) Colonies were formed 8 days after irradiation in miR-300 overexpression (P300) or p53 inhibition (si-p53) group and photographed using a phase contrast microscope. The arrows indicate colonies. Scale bar, 100 μm. (F) Quantitation of SA-β-gal staining A549 cells with miR-300 overexpression (P300) or p53 inhibition (si-p53) 8 days after treated with 10 Gy of X-rays. Ctrl, control, cells exposed to IR but no transfection; NC, pre-miRNA negative control; P300, pre-miR-300; si-p53, p53 siRNA. *P < 0.05, ** P < 0.01, compared to Ctrl.
Furthermore, the survival curves of A549 and H446 cells were measured by colony formation assays after exposed to 0, 1, 3 and 5 Gy of X-rays to assess the cellular radiosensitivity. The survival fraction values were similar between P300 group and si-p53 group in A549 cells, and both groups showed a significant higher survival rate compared to the control group at every dose, especially at 5 Gy of X-rays the survival fraction value of P300 or si-p53 group was approximate 3 times as the control group (P = 0.0023 and 0.0044 respectively, ). The colony formation ability presented a similar trend in H446 cells transfected with pre-miR-300 or p53 siRNA as well as in A549 cells (). In addition, apaf1 silencing by siRNA effectively interfered its expression in H1299 cells treated with 5 Gy of X-rays (), the apoptotic rate decreased from 15.12 ± 1.44% in 5 Gy group to 8.25 ± 1.26% in 5 Gy si-apaf1 group or 8.38 ± 0.92% in 5 Gy P300 group respectively. Collectively, these data demonstrate that miR-300 regulates the cellular sensitivity to IR through targeting p53 and/or apaf1 in lung cancer cells.
miR-300 suppresses IR-induced senescence in A549 cells
Interestingly, A549 cells presented long-term G2 phase arrest when exposed to 10 Gy of X-rays (), the phenomenon we previously reported in melanoma cell line 92-1 cells was named cell cycle suspension.Citation35 However, the underlying molecular mechanism is still elusive. As we have demonstrated that miR-300 was effective to overcome the G2 phase block induced by 2 or 5 Gy of X-rays in A549 cells ( and ), we speculated that miR-300-p53 pathway may be involved in IR-induced cell cycle suspension.
In order to validate the speculation, we first measured the cell cycle distribution in p53 wild type cell lines 92-1, A375 and MCF7 and p53 mutant cell lines 786-O, HeLa and OCM-136 15 or 48 h after treated with 10 Gy of X-rays. The flow cytometry results showed that only p53 wild type cells underwent G2 cell cycle suspension while cells with mutant p53 relieved the G2 block to a great extent at 48h post-irradiation (Fig. S4A and S4B), indicating IR-induced cell cycle suspension is, at least partly, dependent on p53 gene status. On the other hand, we found that ectopic expression of miR-300 greatly released the cells blocked in G2 phase at 48 h after treated with 10 Gy of X-rays in A549 cells ( and ). All the data suggest that miR-300-p53 pathway is to some extent correlated to IR-induced cell cycle suspension.
As cells underwent cell cycle suspension subsequently exhibit senescence but not apoptosis,Citation35 we next studied the role of miR-300 in the cellular responses to p53-related senescence. As presented in , permanent growth arrest was induced by 10 Gy of X-ray irradiation in A549 cells, while overexpression of miR-300 attenuated the cell proliferation inhibition. Moreover, the permanent growth arrest cells showed obvious morphological changes associated with senescence and were barely able to form colonies. However, cells with miR-300 overexpression or p53 silencing were able to form typical colonies ( and S5A). Further, the proportion of SA-β-gal positive cells, a widely used marker for indication of senescence,Citation37,38 was significantly lower when miR-300 was overexpressed or p53 was knocked down ( and S5B). Together, these results suggest that miR-300 enhances the cellular resistance to IR by reducing p53-dependent cell cycle suspension and senescence in A549 cells.
Discussion
Our previous study showed that 53BP1 was a potential target gene of miR-300.Citation15 In this study, we reveal that overexpression of miR-300 elevates the DSBs clearance rate in A549 cells exposed to 0.5 Gy of X-rays although there is no difference in the number of 53BP1 between NC group and P300 group at 1 h post-irradiation, which is inconsistent with previous report,Citation39 indicating that 53BP1 a limited impact of miR-300 on DNA damage repair in A549 cells in low dose radiation. Furthermore, we reveal that miR-300 attenuates the cellular sensitivity to IR via diminishing p53-apaf1-related G2 cell cycle arrest and apoptosis. In particular, miR-300 counteracts IR-induced cell cycle suspension and senescence through suppression of p53 and p53-related pathways in high dose radiation.
In the past few years, several studies have revealed the function of miR-300 in tumor progression, epithelia to mesenchymal transition (EMT) and stemness maintenance of tumor stem cells. Zhang and colleagues found that miR-300 was commonly upregulated in glioma and glioma stem like cells (GSLCs), overexpression of miR-300 could enhance the self-renewal and reduce differentiation of GSLCs through targeting LZTS2.Citation40 Xue et al identified that ectopic expression of miR-300 promoted proliferation, invasion and EMT in osteosarcoma.Citation41 In contrast, Yu et al found that miR-300 negatively regulated EMT by direct targeting Twist and inhibited cancer cell invasion and metastasis.Citation42 Zhou et al demonstrated a downregulated expression of miR-300 in glioblastoma and a tumor suppressor role of miR-300 via inhibiting ROCK1.Citation43 These conflicting results from different reports suggest that miR-300 performs radically opposite functions depending on the cellular context. In the present study, we tested the cell proliferation and cell cycle distribution influenced by ectopic expression of miR-300 in A549 cells and found no effect on the proliferation and the cell cycle under miR-300 overexpression, supposing other regulating pathways result in destabilization or efficient inhibition of p53.
As a transcription factor p53 can activate the transcription of a group of miRNAs, such as miR-34,Citation44 miR-200,Citation45 miR-145 and miR-139-5p,Citation46,47 to protect against tumorigenesis and progression. On the other hand, a series of miRNAs have been validated to suppress the expression and function of p53 through direct or indirect repression of p53. For example, miR-504Citation48 and miR-125bCitation21 impairs p53-mediated apoptosis and cell cycle arrest during the stress response. Excitingly, a recent study reveal that miR-300 is involved in a p53-PTTG1 targeting miRNAs-PTTG1 feedback loop.Citation49 Liang and colleagues identified that DNA damage-induced p53 expression activated miR-300 transcription by directly binding to its promoter while miR-300 indirectly modulated p53 expression by targeting PTTG1 in GH3 cells. In this study, we demonstrated that miR-300 suppressed p53 protein expression through directly binding to p53 mRNA 3′-UTR ( and ).
We confirmed that high dose IR-induced prolonged G2 cell cycle arrest depended on p53 status (Fig. S4A and S4B) and subsequently underwent senescence (Fig. S5B), which is consistent with the previous studies.Citation32,50 However, the mechanism by which sustained G2 arrest triggers senescence is still blurred. In a recent publication, Ye et al revealed that a slippage of long-term G2 arrest into G1 phase was involved in IR-induce senescence, in which cells presented tetraploid karyotypes but with robust activation of G1 phase-specific proteins and inactivation of G2 phase-specific proteins.Citation51 Zhang et al demonstrated that the bypass of mitosis involved in the slippage of long-term G2 arrest into G1 phase was dependent upon p21.Citation52 In addition, they also revealed that the complexity and maintenance of telomeric DNA damage contributed effectively to push the long-term G2 arrested cells into senescence.Citation36 Here, for the first time we report that miRNA is involved in mediation of DNA damage-induced sustained G2 arrest, which lead to a new strategy to explore the mechanism of senescence associated with sustained G2 arrest.
A pervious study demonstrated that miR-300 could delay replicative senescence in cardiac progenitor cells.Citation53 In our experiment, a significant reduction of senescence was observed in miR-300 over-expressing cells treated with high dose IR ( and S5B), suggesting contribution of miR-300 to DNA damage-induced senescence. Interestingly, p53 interference by siRNA or miR-300 overexpression enhanced the cellular resistance to IR. However, p53 inhibition lead to the entrance to M phase from G2 phase block (data not shown) and subsequently underwent apoptosis while this phenomenon was not observed in miR-300 overexpression cells (). As the level and maintenance of p53 play an important part in cell destiny,Citation19 miR-300 not only declines p53 expression, but also suppresses apoptosis by regulating direct or indirect apoptosis-related targets in the context of cellular response to IR. We indeed found that apoptosis and autotrophy-induced apoptosis signaling pathway members,Citation54 such as apaf1, atg2b, atg10 and atg14, were also potential targets of miR-300. Apaf1 is a transcriptional target of p53 and miRNAs can induce degradation of target mRNA,Citation34,55 indicating that the suppression of apaf1 can be directly induced by miR-300 targeting or indirectly by miR-300-mediated p53 silencing. Taken together, miR-300 is involved in regulation of cellular radiosensitivity in both p53 wild type and mutant lung cancer cells.
In summary, our data elucidate a new role of miR-300 as a modulator of radiosensitivity through targeting p53 and apaf1 in lung cancer cells. Our work enriches the miRNA-p53-apaf1 regulatory network in response to IR-induced DNA damage, which provides a novel insight into the role of miRNAs in sensitizing tumor cells to radiotherapy.
Materials and methods
Cell lines
Human non-small cell lung cancer cell line A549, kidney cancer cell line 786-O, breast cancer cell line MCF7 and melanoma cell line A375 were purchased from the Shanghai Institute for Biological Sciences (Shanghai, China). Human non-small cell lung cancer cell line GLC82 and small cell lung cancer cell line NCI-H446 (H446) were a kind gift from Dr. Jianxiu Ma (School of Medicine, Northwest University for Nationalities, Lanzhou, China), NCI-H1299 (H1299) by Dr. Wensen Jin (Teaching and Research Section of Nuclear Medicine, Anhui Medical University, Hefei, China) and human melanoma cell lines 92-1 and OCM-1 by Dr. Guangming Zhou (School of Radiation Medicine and Protection, Soochow University, Suzhou, China). The A549 cells were cultured in DMEM/F-12 medium (Gibco, 12500-062, USA), H1299 cells in DMEM medium (Gibco, 12800-082) and other cell lines in RPMI-1640 medium (Gibco, 31800-105), supplemented with 10% fetal bovine serum (Gibco, 10099-141), 100 units/ml penicillin and 100 mg/ml streptomycin. Cells were maintained at 37˚C in a humidified atmosphere of 95% air and 5% CO2.
Cell transfection and irradiation
2 × 105 cells were seeded in 35mm dishes and cultured for 24 h prior to be transfected with miRNA mimics (Ambion, AM17100 and AM17110, USA) including miR-300 precursor (P300) and the precursor negative control (NC), miR-300 inhibitor (I300) and miRNA inhibitor negative control (IN) or siRNA including p53 siRNA (si-p53, RiboBio, stQ0019919-1, China), apaf1 siRNA (si-apaf1, RiboBio, stQ0003724-1) and the negative control (si-NC, RiboBio, siN05815122147-1-5) at 40-60% confluence using Lipofectamine™ 2000 (Invitrogen, 11668-019, USA). The final concentration of the miRNA mimics was 30 nM and the siRNA was 50 nM. The medium was replaced with fresh culture medium 5 h following transfection. After that, cells were irradiated with a laboratory RX-650 X-ray source (Faxitron, Tucson, Arizona, USA) at a dose rate of 0.8 Gy/min (100 keV, 5mA).
Immunofluorescence assay
Immunoflorescence of 53BP1 was carried out as previously reported.Citation35 In brief, cells were fixed in 4% paraformaldehyde for 10 min and in methanol at ¯20˚C for 20 min, permeabilized in PBS with 0.5% Triton X¯100 for 10 min, blocked with 5% skim milk for 2 h and stained with the primary antibody rabbit anti¯53BP1 antibody (Abcam, ab36823, UK) for 2 h at room temperature. The bound antibody was visualized using Alexa Fluor® 594 anti¯rabbit antibody (Molecular Probes, A-11037, USA), and cell nuclei were counterstained with DAPI solution (Invitrogen, P36941). Images were captured and 53BP1 foci were counted using a fluorescence microscope (Leica, Wetzlar, Hesse-Darmstadt, Germany), and a minimum of 100 cells were scored for each sample.
Flow cytometry assay
Cells transfected with miRNA mimics or siRNA were exposed to X-rays. For cell cycle assay, cells were fixed in -20 ˚C pre-chilled 70% alcohol overnight, and then stained with 20 μg/ml propidium iodide (PI) at room temperature. For apoptosis assay, cells were harvested 24 h post-irradiation and stained with Annexin V-FITC/PtdIns (BD Biosciences, 556547, USA) for 30 min at room temperature. Flow cytometry assay was performed using an Amnis imaging flow cytometer (Merck Millipore, Darmstadt, Hesse, Germany) and at least 10,000 gated events were acquired from each sample. The total apoptotic rate was evaluated as the sum of the early and the late apoptotic rate. The data were analyzed with IDEAS Application v6.0 (Amnis) or FlowJo v6.0 (Tree Star, Ashland, OR, USA).
qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, 15596018). Reverse transcription of miRNA or mRNA was performed with All-in-One™ miRNA First-Strand cDNA Synthesis kit (GeneCopoeia, AMRT-0060, China) or All-in-One™ First-Strand cDNA Synthesis kit (GeneCopoeia, AORT-0060). To quantify the miR-300 expression or mRNA expression of p21 and apaf1, real-time PCR was performed using All-in-One™ miRNA qPCR kit (GeneCopoeia, AMPR-0200) or All-in-One™ mRNA Detection kit (GeneCopoeia, AOPR-0200) based on SYBR-Green. snRNA U6 and GAPDH were used as control for normalization respectively. All PCR primers were purchased from GeneCopoeia (U6, HmiRQP9001; miR-300, HmiRQP0377; GAPDH, HQP006940; p21, HQP071930; apaf1, HQP008934). The relative expression was calculated using the 2-ΔΔCt method.Citation56
Colony formation assay
Cells were harvested by trypsinization and resuspended in DMEM/F-12 or RPMI-1640 medium supplemented with 10% FBS. An appropriate number of cells were plated into a 60 mm dish to produce ∼50-150 colonies. Following incubation for 12 days, cells were fixed with 75% ethanol and stained with 0.5% crystal violet at room temperature. Colonies containing more than 50 cells were counted as survivors, at least three parallel dishes were scored for each treatment and each experiment was conducted at least three times independently.
Western blot
Protein lysates were obtained using RIPA buffer (Beyotime, P0013C, China). Proteins were separated by 10% SDS-PAGE and transferred to a methanol-activated PVDF membrane (Millipore, IPVH00010, USA). The membrane was blocked with 5% skim milk in PBS containing 0.1% Tween-20 for 1 h and subsequently probed with anti-p53 antibody (Abcam, ab131442), anti-apaf1 antibody (Abcam, ab32372), anti-p21 antibody (Abcam, 109199) and anti-gapdh antibody (Abcam, ab9485) for 2 h at room temperature. After 1 h incubation with goat-anti-rabbit HRP-conjugated secondary antibody (Abcam, ab97051), the protein bands were detected with luminal reagent (Millipore, WBKLS0500, USA).
Luciferase reporter assay
The wild type and mutant P53/APAF1-3′-UTR containing the binding sites of miR-300 were synthesized by Sangon Biotech (Shanghai, China). The segments were inserted into pmirGLO Dual-Luciferase miRNA Target Expression Vectors (Promega, E1330, USA). 1 × 105 A549 cells were seeded in 12-well plates for 24 h and co-transfected with 300 ng luciferase vector and miRNA mimics (30 nM final). Dual luciferase assays were performed 24 h after transfection using Dual Luciferase Reporter Assay Kit (Promega, E1910). Renilla luciferase activity was used to normalize the firefly luciferase activity.
Cell growth curves
1 × 105 A549 cells were plated in 12-well plates a day before transfected with miRNA mimics. Cells were exposed to 10 Gy of X-rays after transfection and counted every 4 days using a Coulter Counter (Beckman, Brea, CA, USA).
SA-β-gal staining
1 × 105 A549 cells were plated in 35 mm dishes and cultured for 24 h before transfection with RNA mimics or siRNAs. Then cells were exposed to 10 Gy of X-rays after transfection. Cells were stained by Senescence Associated (SA) β-Galactosidase Staining Kit (Beyotime, C0602) following the protocol 8 d after irradiation. In brief, cells were washed with PBS and fixed for 10 min at room temperature, washed with washing buffer thrice and incubated at 37 ˚C for 24 h with staining solution. Senescent cells were identified and calculated using a light microscope.
Statistical analysis
All the experiments were repeated at least three times and results are shown as mean ± SD. The statistical significance of the results was determined by Student's t-test and *P < 0.05 was considered as a statistically significant difference.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KCCY_S_1367070.pptx
Download MS Power Point (1.5 MB)Acknowledgments
The authors thank Dr. Guangming Zhou, Dr. Jianxiu Ma and Dr. Wensen Jin for their kind help of the cell lines.
Funding
This work was supported by the National Nature Science Foundation of China (NO. 11405232, 31270895 and 31400723), the Science and Technology Research Project of Gansu Province (NO. 145RTSA012) and the Natural Science Foundation of Gansu Province (NO. 145RJZA117).
References
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32. doi:10.3322/caac.21338
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-917. doi:10.1002/ijc.25516. PMID:21351269
- Pignon JP, Arriagada R, Ihde DC, Johnson DH, Perry MC, Souhami RL, Brodin O, Joss RA, Kies MS, Lebeau B, et al. A meta-analysis of thoracic radiotherapy for small-cell lung cancer. N Engl J Med. 1992;327:1618-24. doi:10.1056/NEJM199212033272302. PMID:1331787
- Czochor JR, Glazer PM. microRNAs in cancer cell response to ionizing radiation. Antioxid Redox Signal. 2014;21:293-312. doi:10.1089/ars.2013.5718. PMID:24206455
- Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr Opin Genet Dev. 2013;23:12-9. doi:10.1016/j.gde.2013.01.002. PMID:23453900
- Garofalo M, Croce CM. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist Updat. 2013;16:47-59. doi:10.1016/j.drup.2013.05.001. PMID:23757365
- Hong L, Yang Z, Ma J, Fan D. Function of miRNA in controlling drug resistance of human cancers. Curr Drug Targets. 2013;14:1118-27. doi:10.2174/13894501113149990183. PMID:23834156
- Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics. 2012;10:246-53. doi:10.1016/j.gpb.2012.07.005. PMID:23200134
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389-402. doi:10.1038/nrc2867. PMID:20495573
- Wan G, Mathur R, Hu X, Zhang X, Lu X. miRNA response to DNA damage. Trends Biochem Sci. 2011;36:478-84. doi:10.1016/j.tibs.2011.06.002. PMID:21741842
- He M, Zhou W, Li C, Guo M. MicroRNAs, DNA damage response, and cancer treatment. Int J Mol Sci. 2016;17:E2087. doi:10.3390/ijms17122087. PMID:27973455
- Wu CW, Dong YJ, Liang QY, He XQ, Ng SS, Chan FK, Sung JJ, Yu J. MicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer. PLoS One. 2013;8:e57036. doi:10.1371/journal.pone.0057036. PMID:23437304
- Song L, Lin C, Wu Z, Gong H, Zeng Y, Wu J, Li M, Li J. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS One. 2011;6:e25454. doi:10.1371/journal.pone.0025454. PMID:21980462
- Wang J, He J, Su F, Ding N, Hu W, Yao B, Wang W, Zhou G. Repression of ATR pathway by miR-185 enhances radiation-induced apoptosis and proliferation inhibition. Cell Death Dis. 2013;4:e699. doi:10.1038/cddis.2013.227. PMID:23807228
- He J, Hua J, Ding N, Xu S, Sun R, Zhou G, Xie X, Wang J. Modulation of microRNAs by ionizing radiation in human gastric cancer. Oncol Rep. 2014;32:787-93. PMID:24919435
- Kumar S, Kumar A, Shah PP, Rai SN, Panguluri SK, Kakar SS. MicroRNA signature of cis-platin resistant vs. cis-platin sensitive ovarian cancer cell lines. J Ovarian Res. 2011;4:17. doi:10.1186/1757-2215-4-17. PMID:21939554
- Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431-42. doi:10.1038/sj.cdd.4401183. PMID:12719720
- Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35:672-88. doi:10.1002/humu.22552. PMID:24665023
- El-Deiry WS. The role of p53 in chemosensitivity and radiosensitivity. Oncogene. 2003;22:7486-95. doi:10.1038/sj.onc.1206949. PMID:14576853
- Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44-55. doi:10.1038/nrm1546. PMID:15688066
- Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862-76. doi:10.1101/gad.1767609. PMID:19293287
- Liu Y, Xing R, Zhang X, Dong W, Zhang J, Yan Z, Li W, Cui J, Lu Y. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA Repair (Amst). 2013;12:741-50. doi:10.1016/j.dnarep.2013.06.002. PMID:23835407
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629-34. doi:10.1126/science.2683079. PMID:2683079
- Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435-8. doi:10.1126/science.1076182. PMID:12364621
- Iwabuchi K, Basu BP, Kysela B, Kurihara T, Shibata M, Guan D, Cao Y, Hamada T, Imamura K, Jeggo PA, et al. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J Biol Chem. 2003;278:36487-95. doi:10.1074/jbc.M304066200. PMID:12824158
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7-18. doi:10.1038/nrm3719. PMID:24326623
- Helton ES, Chen X. p53 modulation of the DNA damage response. J Cell Biochem. 2007;100:883-96. doi:10.1002/jcb.21091. PMID:17031865
- Zermati Y, Mouhamad S, Stergiou L, Besse B, Galluzzi L, Boehrer S, Pauleau AL, Rosselli F, D'Amelio M, Amendola R, et al. Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Mol Cell. 2007;28:624-37. doi:10.1016/j.molcel.2007.09.030. PMID:18042457
- O'Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, Scudiero DA, Monks A, Sausville EA, Weinstein JN, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285-300. PMID:9331090
- Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355-8. PMID:15188009
- Mitsudomi T, Steinberg SM, Nau MM, Carbone D, D'Amico D, Bodner S, Oie HK, Linnoila RI, Mulshine JL, Minna JD, et al. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7:171-80. PMID:1311061
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497-501. doi:10.1126/science.282.5393.1497. PMID:9822382
- Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, Muller H, Helin K. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552-8. doi:10.1038/35078527. PMID:11389439
- Robles AI, Bemmels NA, Foraker AB, Harris CC. APAF-1 is a transcriptional target of p53 in DNA damage-induced apoptosis. Cancer Res. 2001;61:6660-4. PMID:11559530
- He J, Li J, Ye C, Zhou L, Zhu J, Wang J, Mizota A, Furusawa Y, Zhou G. Cell cycle suspension: a novel process lurking in G(2) arrest. Cell Cycle. 2011;10:1468-76. doi:10.4161/cc.10.9.15510. PMID:21455017
- Zhang X, Ye C, Sun F, Wei W, Hu B, Wang J. Both Complexity and location of DNA damage contribute to cellular senescence induced by ionizing radiation. PLoS One. 2016;11:e0155725. doi:10.1371/journal.pone.0155725. PMID:27187621
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363-7. doi:10.1073/pnas.92.20.9363. PMID:7568133
- Bassaneze V, Miyakawa AA, Krieger JE. A quantitative chemiluminescent method for studying replicative and stress-induced premature senescence in cell cultures. Anal Biochem. 2008;372:198-203. doi:10.1016/j.ab.2007.08.016. PMID:17920029
- Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, Tewari M, Furnari FB, Taniguchi T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res. 2011;9:1100-11. doi:10.1158/1541-7786.MCR-11-0007. PMID:21693595
- Zhang D, Yang G, Chen X, Li C, Wang L, Liu Y, Han D, Liu H, Hou X, Zhang W, et al. mir-300 promotes self-renewal and inhibits the differentiation of glioma stem-like cells. J Mol Neurosci. 2014;53:637-44. doi:10.1007/s12031-014-0230-x. PMID:24464870
- Xue Z, Zhao J, Niu L, An G, Guo Y, Ni L. Up-Regulation of MiR-300 Promotes Proliferation and Invasion of Osteosarcoma by Targeting BRD7. PLoS One. 2015;10:e0127682. doi:10.1371/journal.pone.0127682. PMID:26010572
- Yu J, Xie F, Bao X, Chen W, Xu Q. miR-300 inhibits epithelial to mesenchymal transition and metastasis by targeting Twist in human epithelial cancer. Mol Cancer. 2014;13:121. doi:10.1186/1476-4598-13-121. PMID:24885626
- Zhou F, Li Y, Hao Z, Liu X, Chen L, Cao Y, Liang Z, Yuan F, Liu J, Wang J, et al. MicroRNA-300 inhibited glioblastoma progression through ROCK1. Oncotarget. 2016;7(24):36529-36538. doi:10.18632/oncotarget.9068. PMID:27145462
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298-307. doi:10.1016/j.cub.2007.06.068. PMID:17656095
- Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875-83. doi:10.1084/jem.20110235. PMID:21518799
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106:3207-12. doi:10.1073/pnas.0808042106. PMID:19202062
- Cao B, Wang K, Liao JM, Zhou X, Liao P, Zeng SX, He M, Chen L, He Y, Li W, et al. Inactivation of oncogenic cAMP-specific phosphodiesterase 4D by miR-139-5p in response to p53 activation. Elife. 2016;5:e15978. doi:10.7554/eLife.15978. PMID:27383270
- Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, Tang LH, Levine AJ, Feng Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell. 2010;38:689-99. doi:10.1016/j.molcel.2010.05.027. PMID:20542001
- Liang HQ, Wang RJ, Diao CF, Li JW, Su JL, Zhang S. The PTTG1-targeting miRNAs miR-329, miR-300, miR-381, and miR-655 inhibit pituitary tumor cell tumorigenesis and are involved in a p53/PTTG1 regulation feedback loop. Oncotarget. 2015;6:29413-27. doi:10.18632/oncotarget.5003. PMID:26320179
- Wiebusch L, Hagemeier C. p53- and p21-dependent premature APC/C-Cdh1 activation in G2 is part of the long-term response to genotoxic stress. Oncogene. 2010;29:3477-89. doi:10.1038/onc.2010.99. PMID:20383190
- Ye C, Zhang X, Wan J, Chang L, Hu W, Bing Z, Zhang S, Li J, He J, Wang J, et al. Radiation-induced cellular senescence results from a slippage of long-term G2 arrested cells into G1 phase. Cell Cycle. 2013;12:1424-32. doi:10.4161/cc.24528. PMID:23574719
- Zhang XR, Liu YA, Sun F, Li H, Lei SW, Wang JF. p21 is Responsible for Ionizing Radiation-induced Bypass of Mitosis. Biomed Environ Sci. 2016;29:484-93. PMID:27554118
- Cruz FM, Tome M, Bernal JA, Bernad A. miR-300 mediates Bmi1 function and regulates differentiation in primitive cardiac progenitors. Cell Death Dis. 2015;6:e1953. doi:10.1038/cddis.2015.255. PMID:26512961
- Gump JM, Thorburn A. Autophagy and apoptosis: what is the connection? Trends Cell Biol. 2011;21:387-92. doi:10.1016/j.tcb.2011.03.007. PMID:21561772
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118-26. doi:10.1016/j.tcb.2006.12.007. PMID:17197185
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-8. doi:10.1006/meth.2001.1262. PMID:11846609
