 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The nucleolar proteins which link cell proliferation to ribosome biogenesis are regarded to be potentially oncogenic. Here, in order to examine the involvement of an evolutionary conserved nucleolar protein SURF6/Rrp14 in proliferation and ribosome biogenesis in mammalian cells, we established stably transfected mouse NIH/3T3 fibroblasts capable of conditional overexpression of the protein. Cell proliferation was monitored in real-time, and various cell cycle parameters were quantified based on flow cytometry, Br-dU-labeling and conventional microscopy data. We show that overexpression of SURF6 accelerates cell proliferation and promotes transition through all cell cycle phases. The most prominent SURF6 pro-proliferative effects include a significant reduction of the population doubling time, from 19.8 ± 0.7 to 16.2 ± 0.5 hours (t-test, p < 0.001), and of the length of cell division cycle, from 17.6 ± 0.6 to 14.0 ± 0.4 hours (t-test, p < 0.001). The later was due to the shortening of all cell cycle phases but the length of G1 period was reduced most, from 5.7 ± 0.4 to 3.8 ± 0.3 hours, or by ∼30%, (t-test, p < 0.05). By Northern blots and qRT-PCR, we further showed that the acceleration of cell proliferation was concomitant with an accumulation of rRNA species along both ribosomal subunit maturation pathways. It is evident, therefore, that like the yeast homologue Rrp14, mammalian SURF6 is involved in various steps of rRNA processing during ribosome biogenesis. We concluded that SURF6 is a novel positive regulator of proliferation and G1/S transition in mammals, implicating that SURF6 is a potential oncogenic protein, which can be further studied as a putative target in anti-cancer therapy.
Introduction
Ribosome biogenesis is a complex, time-ordered and energy-consuming process that occurs mainly in the nucleolus. In nucleoli of mammalian cells, rRNA genes are transcribed to yield the primary 47S pre-rRNA transcripts, which then undergo numerous cleavage reactions, post-transcriptional modifications and interactions with appropriate proteins to provide a cell with the required number of 18S, 5.8S and 28S rRNAs and ribosomes.Citation1,Citation2 Ribosome synthesis is strictly coordinated with cell growth and proliferation, so that actively dividing cells, including tumor and embryo cells, have particularly well developed nucleoli.Citation3 In line with these data, in vitro experiments showed that dysregulation of nucleolar proteins either leads to an arrest of cell cycle and drives cells to apoptosis or, in opposite, facilitates cell transformation and uncontrolled proliferation.Citation4,Citation5 Consequently, identification of the nucleolar proteins involved in cell cycle regulation is important not only for elucidating mechanisms which underlie the relationship between nucleolar functions and cell cycle control but also towards the development of novel therapeutic interventions for cancer treatment.Citation5-9
Several auxiliary rDNA transcription regulators and numerous rRNA processing factors such as endo- and exoribonucleases, RNA helicases, NTPases and methyltransferases are involved in the synthesis and maturation of rRNAs.Citation10,Citation11 Some of these factors are multifunctional proteins, which in addition to making ribosomes also participate in cell proliferation. Among these factors are the abundant nucleolar chaperons NPM1 (B23/nucleophosmin) and nucleolin/C23 which promote cell proliferation through stimulatory effects on rDNA transcription, ribosome subunit export and DNA replication.Citation12,Citation13 Overexpression of the Bop1 protein, a member of the PeBoW complex that is essential for maturation of the large ribosomal subunit, impairs cell proliferation by increasing the percentage of abnormal multipolar mitotic figures.Citation14,Citation15 In addition, several nuclear and cytoplasmic proteins with a partial occurrence in nucleoli are also linked to proliferative processes. Such proteins include the nuclear proteins CHD7,Citation16 Las1L,Citation17 DHX33 helicase,Citation18 Sirt6,Citation19 PHF6,Citation20 and NSA2Citation21 as well as ribosomal proteins S3aCitation22 and L36a.Citation23
SURF6 has been described as a nucleolar protein that belongs to an evolutionary conserved protein family, which members are present in fungi, animal and plant kingdoms but are absent in bacteria.Citation24 The characteristic feature of the SURF6 proteins is the presence of a conservative domain positioned in the carboxyl terminus that has an average residue identity of 36% between different species and is enriched with lysine and arginine residues, which confer its overall basic charge (pI > 10).Citation24 In mammals, SURF6 is a product of one of the six genes comprising the surfeit gene locus and is a typical housekeeping protein that is expressed in various tissues and throughout cell cycle in NIH/3T3 fibroblasts.Citation25,Citation26 A particularly high level of SURF6 expression has been described in actively proliferating cells, such as embryonic, progenitor and hematopoietic stem lineagesCitation27 and in some cancer cells (http://www.proteinatlas.org/ENSG00000148296-SURF6/cancer). Interestingly, in mouse spleenocytes activated for proliferation in vitro expression of SURF6 begins before that of the PCNA and Ki-67 proteins, which are the main proliferation markers in normal and cancer cells.Citation28
Presently, the known functions of SURF6 have been elucidated mainly from studies on budding yeasts, where its homologue is named Rrp14/ykl082c.Citation29,Citation30 Rrp14p has been shown to play multiple roles in ribosome biogenesis, from synthesis of the primary 35S pre-rRNA transcript to assembly of the large and small ribosomal subunits. Additionally, knockout of Rrp14p retards the yeast proliferation by causing defects in budding and organization of the mitotic spindle. However, specific roles of SURF6 in proliferation and ribosome biogenesis in mammals await additional investigation. Based on large-scale cDNA transfection screening of colony formation, SURF6 has been identified as a putative cancer-related protein in cultured mouse fibroblasts and human cancer cells.Citation31 We have also shown that transient knockdown of SURF6 promotes death in mouse fibroblasts.Citation32 However, the degradation of rRNA known to occur in dead cells Citation33 did not allow us to examine a direct link between proliferation and ribosome production in SURF6-depleted cells.
In this study, in order to clarify the implication of SURF6 in proliferation and ribosome biogenesis, we establish a stable sub-line of mouse NIH/3T3 fibroblasts (called NIH/3T3-174 fibroblasts) which are capable to overexpress SURF6 conditionally in the presence of doxycycline. We show that upon appropriate doxycycline concentrations induction of SURF6 overexpression has no notable cytotoxicity but significantly accelerates proliferation unless the induced fibroblasts reach cell-cell contacts. Like its yeast homologue Rrp14, mammalian SURF6 is involved in rRNA processing along both ribosomal subunit maturation pathways. Overall, our data demonstrate that mammalian SURF6 is an rRNA processing factor, which promotes proliferation and accelerates G1/S transition in non-malignant fibroblasts. Our data endorse the hypothesis that mammalian SURF6 is a putative oncoprotein.Citation31
Results
Phenotype of stably transfected NIH/3T3-174 fibroblasts
In order to determine the effect of the protein SURF6 on the proliferation and ribosome biogenesis in mammalian cells, we establish a stable sub-line of mouse NIH/3T3 fibroblasts (called NIH/3T3-174 fibroblasts), which are capable to overexpress SURF6 in the presence of doxycycline. In , the plasmid construct used to obtain stably transfected NIH/3T3-174 fibroblasts is shown. On Western blots of control (-Dox) and induced (+Dox) cells SURF6 is visible as a major band with an apparent molecular mass of 43 kDa that corresponds to the electrophoretic mobility of mouse SURF protein Citation25 (). After 24 hours of 100 ng/ml doxycycline administration, the amount of SURF6 becomes about three times and after 48 hours – up to10 times higher than in -Dox cells. A weaker and more mobile band present in SURF6-overexpressing cells results most probably from a partial degradation of the protein. According to qRT-PCR results obtained in different experiments, the number of SURF6 mRNA copies increased from 2.5-3 (at 24 hrs of post-induction) to 6–8 (at 48 hrs) times (data not shown).
Figure 1. (A) A diagram of the pBI-SURF6 plasmid used for generation of stably transfected mouse NIH/3T3-174 fibroblasts capable to overexpress SURF6 in the presence of doxycycline (Dox). ampR – ampicillin-resistence gene, EGFP – the sequence coding for the EGFP protein, CMV – minimal CMV promoter, TRE – tetracycline-responsive element, SURF6 cDNA – cDNA of the mouse SURF6 gene, Surf6. (B) Immunoblots of control and induced NIH/3T3-174 fibroblasts after incubation without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 and 48 hours. SURF6 was revealed with the anti-SURF6 serum,Citation26 β-tubulin (a loading control) was recognized with a commercial antibody. Star (*) indicates a position of a faster migrating SURF6 form appeared in +Dox fibroblasts. (C-J) General images of NIH/3T3-174 fibroblasts cultured without (-Dox; c, d) or with (+Dox; E-J) 100 ng/ml doxycycline for 48 (C-F and J) or 24 (G-I) hours. (C, E, and H) – EGFP fluorescence, (D and F) – phase contrast, (G and J) – immunolabeling for SURF6; (I) – DAPI staining of chromatin. In the absence of doxycycline, almost no EGFP fluorescence is seen (C). In the presence of doxycycline, the cells are intensely marked for EGFP (E and H), hereby indicating an effectiveness of the pBI-SURF6 plasmid. The phenotype of +Dox fibroblasts (F) remains similar to that of -Dox cells (D). SURF6 is distributed in nucleoli (G and J). A less intense anti-SURF6 immunolabeling of nucleoli is generally seen in cells with a weaker EGFP fluorescence (asterisks, G and H). Large-scale nuclear organization looks similar in different cells (I). Scale bars, (C-F) – 100 µm, (G-J) – 30 µm. (K) Trypan blue excision assay. Bar graphs illustrating the number of dead cells in –Dox (control, grey columns) and +Dox (black columns) cells after 24 and 48 hours of the induction. At each time-point, the control and experimental values do not differ statistically significant (p > 0.05). The data are shown as the mean ± SEM (n = 3).
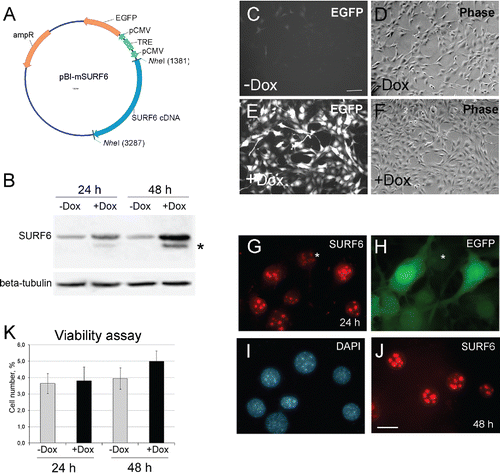
In , representative microscopic images of NIH/3T3-174 fibroblasts incubated without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 48 hours are shown. The control (-Dox) fibroblasts ( and ) are almost EGFP-negative (), whereas the majority of +Dox cells ( and ) are brightly fluorescent (). Like in non-transfected cells, Citation25,Citation26 in +Dox NIH/3T3-174 fibroblasts, SURF6 distributes mainly within nucleoli ( and ). It is worth mentioning that in cells with a stronger EGFP fluorescence, nucleoli are more intensely stained for SURF6 than the nucleoli in weakly EGFP-fluorescent cells ( and , asterisks). There were no visible changes in large-scale chromatin configuration upon SURF6 overexpression (). We also did not observe significant difference in the number of dead cells in -Dox and +Dox cell populations neither by conventional microscopy ( and ) nor by the trypan blue excision test (). These data support a conclusion that excess of SURF6 does not cause significant cytotoxic effects at least within 24–48 hours of the induction.
Effects of SURF6 overexpression on cell proliferation
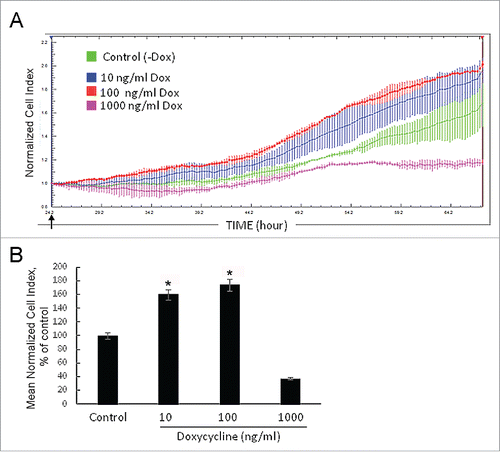
First, we examined effects of the SURF6 excess on cell proliferation in real time using the xCELLigence (RTCA)-DP system. Experiments were repeated several times, and the results of a representative experiment are shown ( and ). The proliferation rate of NIH/3T3-174 fibroblasts was monitored in the presence of 10 ng/ml, 100 ng/ml or 1000 ng/ml doxycycline simultaneously with that of non-induced (-Dox) cells (). In , there are bar graphs illustrating the results of quantification of the curve slopes between 24 hours (the time-point of the doxycycline administration) and ∼70 hours (the experiment terminus). Both images demonstrate that 1000 ng/ml doxycycline appeared to suppress an increase of the cell number. On the contrary, 10 ng/ml and 100 ng/ml doxycycline start to promote cell proliferation shortly (∼5-6 hrs) after the induction. The stimulating effects were intensified with time so that after 24–30 hours of the treatment the differences between the normalized cell indexes in the induced and control cells became significant (p < 0.05; and ). Therefore, we concluded that induction of SURF6 overexpression observed in the presence of both doxycycline concentrations (i.e., 10 ng/ml and 100 ng/ml) confers fibroblasts with a proliferative advantage. Taking into account that doxycycline can be instable in neutral aqueous solutions, including culture medium, the antibiotic concentration of 100 ng/ml was used in all further experiments.
Figure 2. Growth rate of NIH/3T3-174 fibroblasts monitored in real time with the xCELLigence (RTCA)-DP instrument system. (A) Cells (∼3,000/well) were seeded in duplicate in an E-16 plate and cultured in the absence (-Dox, control) or the presence of 10 ng/ml, 100 ng/ml and 1000 ng/ml (+Dox) doxycycline. Doxycycline in DMEM (+Dox) or the equal volume of DMEM alone (-Dox) were added after 24 hours of cell seeding (arrow). The horizontal axis – time in hours from the beginning of the experiment. The vertical axis – the normalized cell index automatically registered every 15 min. Small vertical bars in the curves are fluctuations of the index at each registration time-point occurred in duplicates. Doxycycline in the concentration of 1000 ng/ml (pink curve) represses cell growth, while 10 ng/ml (blue curve) and 100 ng/ml (red curve) doxycycline stimulate proliferation as compared with -Dox cells (green curve). The SURF6 pro-proliferative effects start to be visible after 5–6 hours of the induction (corresponds to ∼30 hrs on the horizontal axis) and intensify with time. (B) Bar graphs illustrating the mean normalized cell index in +Dox cells in the percentage to control (-Dox) values considered as 100%. The proliferation rate was determined by analyzing the slopes of each curve obtained between the doxycycline administration (at ∼24 hrs of post-seeding) and the end of an experiment (∼70 hrs of post-seeding; n = 3). The results are expressed as the mean ± SEM. (*) indicate the values which differ insignificantly (p > 0.05). Induction of SURF6 overexpression with 10 ng/ml and 100 ng/ml doxycycline significantly (p < 0.05) activates proliferation as compared with non-stimulated NIH/3T3-174 fibroblasts, whereas 1000 ng/ml doxycycline turns out to be cytotoxic for the cells.
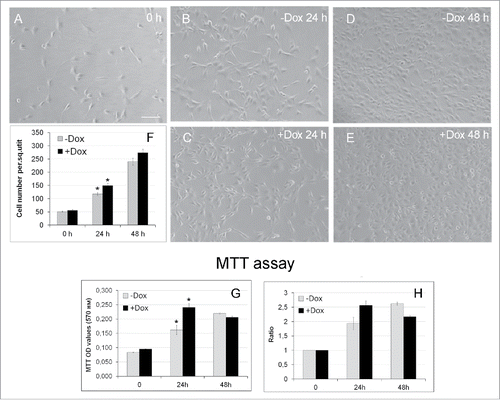
We then examined the proliferation rate of +Dox and –Dox NIH/3T3-174 fibroblasts by monitoring their number over time with conventional phase contrast microscopy and the MTT assay. For the microscopy, fibroblasts were seeded in Petri dishes, and cultured for 3 hours to ensure cell attachment to substrate (the T0 time-point; ). Twenty-five randomly selected fields of view were photographed shortly after and then after 24 and 48 hours cell culturing in the absence ( and , -Dox) or the presence (, +Dox) of doxycycline. The cell number per frame was counted at every time-point in five independent experiments, averaged, and is presented as the mean ± SEM in . The Figure shows that after the induction, the number of +Dox cells/square unit exceeds that in -Dox cells, thereby indicating that the induced cells grow faster than the untreated controls. After 24 hours, the differences between the-Dox and +Dox values are significant (p < 0.05). Similar results were also obtained with the MTT assay that measures cell proliferation and viability. Thus, after 24 hours of the induction the number of viable cells in +Dox samples significantly (p < 0.05) exceeded that in –Dox cells ( and ). Thus, three independent approaches, namely the xCELLigence analysis of cells in real time, conventional microscopy and the MTT assay showed that incubation of NIH/3T3-174 fibroblasts with 100 ng/ml doxycycline stimulated cell proliferation, and that this effect was particularly prominent after ∼24 hours of the induction of SURF6 overexpression.
Figure 3. Effects of SURF6 overexpression on proliferation and viability of NIH/3T3-174 fibroblasts cultured without (-Dox) or with (+Dox) 100 ng/ml doxycycline up to 48 hours as analyzed with conventional phase contrast microscopy (A-F) and the MTT assay (G, H). (A-F) Cells were seeded in Petri dishes, cultured in complete growth medium for 3–4 hours to retrieve cell attachment to substrate. Twenty-five random fields of view were photographed shortly after doxycycline administration (“0” time-point, A) and 24 (B and C) and 48 (D and E) hours later, and representative images of the cells are shown in (A-E). Experiments were repeated five times, and in (F) bar graphs illustrating the mean cell number per field ± SEM are shown. The horizontal axis – time in hours, the vertical axis – the number of cells per square unit, small vertical bars – SEM. (G and H) Bar graphs illustrating MTT assay results. The horizontal axes – time in hours, the vertical axes in panel (G) – OD values at 570 nm, in panel (H) – the ODT /OD0 ratios equal to the OD values scored after 24 and 48 hours and normalized to the relative OD values at the “0” time-point. The data are presented as the mean ± SEM based on the results of three independent experiments.
Calculation of the population doubling time
Taking into account the data mentioned above, we calculated the population doubling time at 24 and 48 hours of post-induction as described in Materials and Methods (see the formula No Equation1(1)
(1) ). The results are summarized in . They show that after the first 24 hours the doubling time was 16.2 ± 0.5 hours in +Dox NIH/3T3-174 fibroblasts and 19.8 ± 0.7 hours in the –Dox fibroblasts, or diminished by ∼ 20% (t-test, p < 0.05). At 48 hours, the doubling time was equal to 26.6 ± 1.5 hours in +Dox NIH/3T3-174 fibroblasts and 21.9 ± 1.9 hours in the –Dox fibroblasts, however, these differences turned out to be statistically insignificant (p > 0.05).
Table 1. Population doubling time (in hours) in different types of NIH/3T3 fibroblasts as calculated with the formula 1 (see Material and Methods for detail) after cell culturing without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 or 48 hours. The data are expressed as the mean ± SEM (Standard Error of the Mean). At 24 hours of post-induction, the values observed in –Dox and +Dox NIH/3T3-174 fibroblasts differ statistically significantly (t-test, p < 0.001) and are shown in bold. The population doubling time in –Dox NIH/3T3-174, -Dox NIH/3T3-EGFP, +Dox NIH/3T3-EGFP and parental NIH/3T3 fibroblasts do not differ significantly (p > 0.05). At 48 hours, the population doubling-time between different fibroblasts differs insignificantly (p > 0.05).
In our NHI/3T3-174 cell model, doxycycline increased the level of SURF6 together with induction of expression of EGFP (), that is a protein which can be toxic for cultured mammalian cells.Citation34,Citation35 To determine whether EGFP affects proliferation in NIH/3T3 fibroblasts, we used stably transfected NIH/3T3-EGFP cells, which in the presence of doxycycline expressed EGFP alone. As an additional control, non-transfected (parental) NIH/3T3 fibroblasts were also used. The cells were cultured with or without 100 ng/ml doxycycline for 24 and 48 hours. The results obtained are included in . They show that after 24 hours, the population doubling time (∼18-19 hrs) in all control cells (i.e., +Dox and -Dox NIH/3T3-EGFP and parental NIH/3T3 fibroblasts) is similar to that of NIH/3T3-174 cells grown without doxycycline (19.8 ± 0.7 hrs). According to the t-test, in these cells the differences in population doubling time was not significant (p > 0.05). We concluded therefore that in our cell system expression of EGFP itself did not statistically significantly influence proliferation of NIH/3T3-174 fibroblasts at least within 24 hours of the induction, and that the diminution of the population doubling time observed in +Dox NIH/3T3-174 cells was due to the increasing level of SURF6 but not to the expression of EGFP. At the 48 hours time-point the population doubling time was 23.3 ± 2.5 hours in -Dox NIH/3T3-EGFP cells, 27.7 ± 5.4 hours in +Dox NIH/3T3-EGFP cells and 24.2 ± 3.2 hours in parental NIH/3T3 fibroblasts. These values did not differ significantly (p > 0.05) because of essential variation (SEM) between cells in all these populations. Thus, in our experimental system the proliferation rate of +Dox and -Dox cells differed statistically significant (p < 0.05) after one day the induction of SURF6 overexpression, i.e. when fibroblasts were not in the confluent state and their proliferation were not suppressed by contact inhibition (). Taking into account these observations, particular cell cycle parameters in +Dox and -Dox NIH/3T3-174 cells were determined in at the 24 hour time-point.
Flow cytometry and timing of cell cycle phases
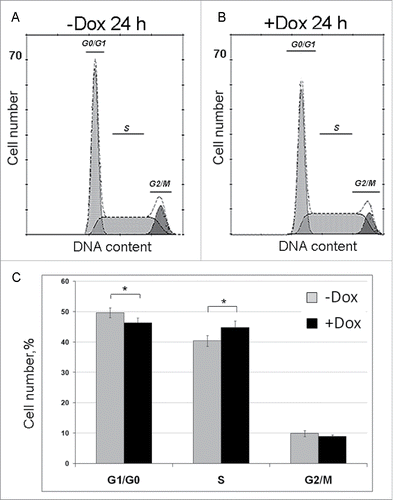
To calculate duration of individual cell cycle phases, we used fluorescence activated flow cytometry. The results of four independent experiments are shown in , where (A) and (B) illustrate one representative experiment, and the panel (C) are the bar graphs illustrating the average number of cells at G0/G1, S and G2/mitosis (G2/M) phases which were calculated taking into account all experimental data. SURF6 overexpression decreases the percentage of cells in G0/G1 phases and simultaneously increases the number of S phase cells. The percentage of G2/M cells do not change notably. These data indicate that an excess of SURF6 affects mainly the number of G0/G1 and S cells.
Figure 4. Effects of SURF6 overexpression on cell cycle progression in NIH/3T3-174 fibroblasts as examined by flow cytometry after 24 hours of post-induction with 100 ng/ml doxycycline. (A and B) Results of a representative experiment, where cells were cultured without (-Dox, A) or with (+Dox, B) doxycycline. In the panel (C), bar graphs illustrate the mean percentage of +Dox (black columns) and -Dox (grey columns) cells which was calculated on the basis of four independent experiments. In G0/G1 phase the number of +Dox cells (46.2 ± 2.0%) is significantly lower (p < 0.05) that the number of -Dox cells (49.6 ± 1.9%), while in S phase the number of +Dox cells (44.9 ± 2.6%) significantly (p < 0.05) exceeds that of -Dox cells (40.5 ± 2.0%). The numbers of +Dox and –Dox cells at G2/mitosis are rather similar. Data are presented as the mean ± SEM.
The flow cytometry data were then used to calculate the duration of cell cycle phases according to the formulae No Equation2(1)
(1) -Equation5
(5)
(5) (see Materials and Methods for detail). The results are summarized in . The Table shows that in +Dox cells, the cell cycle length (14.0 ± 0.4 hrs) was essentially less (t-test, p < 0.001) than in –Dox cells (17.6 ± 0.6 hrs), or was reduced by ∼ 25%. The cell cycle was shortened due to the shortening of all its phases but only G1 phase was reduced significantly (p < 0.05) from 5.7 ± 0.4 hours in –Dox cells to 3.8 ± 0.3 hours in +Dox cells. The durations of S and G2/mitosis phases decreased slightly and insignificantly (p > 0.05; ).
Table 2. Duration (in hours) of cell cycle phases in sub-confluent NIH/3T3-174 fibroblasts cultured without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 hours after seeding. In brackets, the formulae used for calculations are indicated (see Materials and Methods for detail). The percentage of cells at G0/G1, S and G2/mitosis phases was determined by flow cytometry. The percentage of G0 cells was measured by cell population culturing with Br-dU for 24 hours. The number of G1 cells was as the number of G1/G0 cells minus the number of G0 cells. Data are expressed as the mean ± SEM. The compared pairs of values, which according to the t-test, differ significantly (p < 0.05) are indicated in bold. The duration of the cell division cycle in -Dox cells (17.6 ± 0.6) and in +Dox cells (14.0 ± 0.4) differ with p < 0.001.
We also noticed that overexpression of SURF6 increased the mitotic index from 4.4 ± 0.4% in -Dox cells to 5.7 ± 1.3% in +Dox cells, but these values did not differ significantly (p > 0.05).
Putative mechanisms of SURF6 pro-proliferative effects
Like in normal cells,Citation25,Citation26 in +Dox NIH/3T3-174 fibroblasts SURF6 is located in nucleoli ( and ). Considering this, we decided to examine whether SURF6 pro-proliferating effects are linked to ribosome biogenesis. To reach the aim, RNA was isolated from fibroblasts cultured without or with 100 ng/ml doxycycline for 24 hours, and then analyzed by Northern blots with [32P]-labeled probes targeting the 18S rRNA or ITS2 regions of the 47S pre-rRNA transcript. In , the map of the 47S pre-rRNA (A) and representative blots obtained two independent experiment (B and C) are shown. illustrates the average coefficients (black columns) calculated by normalization of the intensity values of each hybridization band observed +Dox cells to the corresponding values in -Dox cells, which are considered as equal to 1 (grey columns). The coefficients were calculated based on the results of several experiments. SURF6 overexpression noticeably increases the amount of all analyzed rRNA intermediates except for 36S rRNA () that is the longest common precursor for 5.8S and 28S rRNAs (). The most prominent changes concern accumulation of 45S pre-rRNA (a common precursor for 18S, 5.8S and 28S rRNA formed downstream from 47S pre-rRNA), 34S and 20S rRNAs (the precursors for 18S rRNA) and of 32S rRNA that is formed along the 28S rRNA maturation pathway downstream from 36S rRNA ().
Figure 5. The diagram of the primary 47S pre-rRNA in the mouse (A), Northern blot (B, C, and E) and qRT-PCR (F) analysis of rRNA expression in NIH/3T3-174 fibroblasts cultured without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 hours. (A) 5′ETS, the 5′ external transcribed spacer; ITS1 and ITS2, the internal transcribed spacers 1 and 2 correspondingly; 18S, 5.8S and 28S are 18S, 5.8S and 28S rRNAs correspondingly. Small vertical bars and designations above the schemes indicate positions of the known pre-rRNA cleavage sites. Small horizontal bars above the schemes indicate positions of the PCR amplicons for 5′ETS and 18 S, 5.8 S, 28 S rRNA (according to ref. Citation1). (B and C) RNA was isolated from sub-confluent fibroblasts and hybridized with [P32] labeled probes recognizing the 18S rRNA (B) or ITS2 (C) regions of the 47S pre-rRNA transcript (A). Arrows indicate positions of the main rRNA species. (D) In mouse cells, processing of the 45S pre-rRNA proceeds mainly through two alternative pathways. In pathway 1, the pre-rRNA processing is initiated in the 5′-ETS to yield 41S pre-rRNA, while in pathway 2, the first cleavage occurs in ITS1 at site 2c to yield 34S rRNA (the longest precursor for 18S rRNA) and 36S rRNA (the longest precursor for 28S rRNA). The 41S pre-rRNA is further cleaved at site 2c into 20S and 36S rRNAs, which are then processed to 18S and 28S rRNAs correspondingly. (according to Citation1). (E) Bar graphs illustrating intensities of rRNA hybridization signals in +Dox cells (black columns) normalized to the values in -Dox cells (grey columns). Data in +Dox cells are presented as the mean ± SEM based on quantification of the results of four Northern blots. (F) Bar graphs illustrates the amount of the 47S pre-rRNA (the 5′ETS amplicons), 18S, 5.8S and 28S rRNAs in +Dox cells (black columns) normalized to the values in -Dox cells (grey columns). The values obtained in +Dox cells are presented as the mean N ± SEM based on the results of five independent experiments.
![Figure 5. The diagram of the primary 47S pre-rRNA in the mouse (A), Northern blot (B, C, and E) and qRT-PCR (F) analysis of rRNA expression in NIH/3T3-174 fibroblasts cultured without (-Dox) or with (+Dox) 100 ng/ml doxycycline for 24 hours. (A) 5′ETS, the 5′ external transcribed spacer; ITS1 and ITS2, the internal transcribed spacers 1 and 2 correspondingly; 18S, 5.8S and 28S are 18S, 5.8S and 28S rRNAs correspondingly. Small vertical bars and designations above the schemes indicate positions of the known pre-rRNA cleavage sites. Small horizontal bars above the schemes indicate positions of the PCR amplicons for 5′ETS and 18 S, 5.8 S, 28 S rRNA (according to ref. Citation1). (B and C) RNA was isolated from sub-confluent fibroblasts and hybridized with [P32] labeled probes recognizing the 18S rRNA (B) or ITS2 (C) regions of the 47S pre-rRNA transcript (A). Arrows indicate positions of the main rRNA species. (D) In mouse cells, processing of the 45S pre-rRNA proceeds mainly through two alternative pathways. In pathway 1, the pre-rRNA processing is initiated in the 5′-ETS to yield 41S pre-rRNA, while in pathway 2, the first cleavage occurs in ITS1 at site 2c to yield 34S rRNA (the longest precursor for 18S rRNA) and 36S rRNA (the longest precursor for 28S rRNA). The 41S pre-rRNA is further cleaved at site 2c into 20S and 36S rRNAs, which are then processed to 18S and 28S rRNAs correspondingly. (according to Citation1). (E) Bar graphs illustrating intensities of rRNA hybridization signals in +Dox cells (black columns) normalized to the values in -Dox cells (grey columns). Data in +Dox cells are presented as the mean ± SEM based on quantification of the results of four Northern blots. (F) Bar graphs illustrates the amount of the 47S pre-rRNA (the 5′ETS amplicons), 18S, 5.8S and 28S rRNAs in +Dox cells (black columns) normalized to the values in -Dox cells (grey columns). The values obtained in +Dox cells are presented as the mean N ± SEM based on the results of five independent experiments.](/cms/asset/9fce90ac-d56c-4786-ab06-df89fd18b0e2/kccy_a_1371880_f0005_b.gif)
The data of qRT-PCR analysis of the PCR-products (amplicons) corresponding to the 5′ETS, 18S, 5.8S and 28S rRNA sequences () after 24 hours of the induction are shown in . The black columns are the mean of fold change values (N) determined in +Dox cells and normalized to the mean values in –Dox controls in four independent experiments. Overexpression of SURF6 increases the number of all amplicons up to two times. A higher number of the amplicons corresponding to the short-lived 47S pre-rRNA (detected with the 5′ETS probe) favors the assumption that an enhanced level of SURF6 stimulates rDNA transcription. An increased level of other detected amplicons, in the whole, corresponds to the Northern blot data favoring an accumulation of the rRNA intermediates upon SURF6 overexpression.
Discussion
SURF6 and cell proliferation
A loss-of-function (knockdown) approach is routinely used to examine a putative involvement of nucleolar proteins in cell cycle control. Based on this approach, the main nucleolar rRNA processing factors NPM1/B23/nucleophosmin,Citation36 nucleolin/C23 13 and nucleostemin Citation37,Citation38 have been recognized as positive regulators of cell proliferation in various cells. Similarly, we applied a loss-of-function approach for studying SURF6 in transiently transfected NIH/3T3 fibroblasts and showed that knockdown of SURF6 affected cell cycle progression.Citation32 However, the loss-of-function approaches are usually limited for drawing fine conclusions on the link between proliferation and ribosome biogenesis by nucleolar proteins, such as SURF6, as these proteins are vital and their knockdown causes cell death and rRNA degradation.Citation33 Consequently, a conditional gain-of-function (overexpression) approach can be more appropriate in cell proliferation studies.
Data illustrating a capability of nucleolar proteins to accelerate proliferation upon inducible expression remain strikingly scarce. To the best of our knowledge, only the NPM1 protein,Citation39 a nuclear protein with a partial nucleolar occurrence NSA2/TINP1,Citation21 and ribosomal proteins RPL36aCitation23 and RPS3a Citation22 have been shown to be involved in proliferation by gain-of-function approaches. Experimentally induced overexpression of NSA2 (the Nop seven-associated 2 protein) facilitates transition of G1 phase to S phase in human cancer cells. NPM1, RPL36a and RPS3a stimulate transformation of normal NIH/3T3 fibroblasts by promoting colony formation and anchorage-independent growth in vitro and contribute to tumorgenesis in vivo. However, mechanisms of these phenomena have not been examined. Conditional overexpression of many other nucleolar proteins either suppresses proliferation (e.g., nucleostemin,Citation37 PAK1IP1,Citation40 Bop1Citation14 or does not exert notable effects (WDR12, Pes1 Citation14).
Here, by using stably transfected NIH/3T3-174 fibroblasts capable of conditional overexpression of SURF6 and by several independent approaches, i.e. monitoring cell proliferation in real time with the xCELLigence system, flow cytometry, and conventional phase-contrast microscopy, we have found that overexpression of SURF6 can promote proliferation of normal mammalian cells. In sub-confluent NIH/3T3-174 fibroblasts, overexpression of SURF6 diminishes the population doubling time (by ∼20%, p < 0.001), the duration of cell division cycle (by ∼25%, p < 0.001), and specifically shortens G1 phase (by ∼30%, p < 0.05) as compared with the non-induced cells ( and ). Moreover, calculations of the duration of cell cycle phases showed that SURF6 is unlikely to control S phase rather to be involved in the G1/S transition (). Thus, our results indicate that the nucleolar protein SURF6 plays a role in cell proliferation control as a positive regulator of G1/S transition in non-malignant cells.
Importantly, the SURF6 pro-proliferating effects could be manifested only upon certain experimental conditions. Specifically, 1000 ng/ml doxycycline, the dose recommended for the Tet-On system in mammalian cells,Citation41 appeared to suppress the proliferation in NIH/3T3-174 fibroblasts (). An optimal doxycycline concentration, 100 ng/ml, significantly activated proliferation in sub-confluent fibroblasts but became less effective, when the cells reached confluency. Thus, after 48 hours of the induction, when a monolayer became nearly complete (), the cell proliferation doubling-time in +Dox cells (26.6 ± 1.5 hours) exceeds that in -Dox cells (21.9 ± 1.9 hours) despite the differences between these values are insignificant (p > 0.05) (). Taking into account that proliferation of mouse 3T3 fibroblasts is inhibited when they reach confluence Citation50 and that in HeLa cells SURF6 interacts with the specific RNA polymerase I cofactor UBF,Citation42 which is suppressed in steady-state fibroblasts,Citation43 one could assume that SURF6 pro-proliferation activity may be mediated by cell contact inhibition and UBF.
SURF6 and ribosome biogenesis
The yeast homologue of SURF6, the Rrp14 protein, functions in ribosome biogenesis as a regulatory factor required for maturation of both ribosomal subunits. In addition, in Rrp14p-depleted cells 35S pre-rRNA synthesis appeared to decline.Citation29,Citation30 However, ribosome biogenesis steps, which recruit SURF6 in mammalian cells, remain undefined. Here we show that in mouse fibroblasts conditional overexpression of SURF6 affects various steps of rRNA processing. Based on the Northern blot data, SURF6 overexpression causes accumulation of 34S and 20S rRNAs (the precursors for 18S rRNA) and of 32S rRNA that is a precursor for 28S rRNA (). Nonetheless, we did not observe an accumulation of the longest common precursor for 5.8S and 28S rRNAs, the 36S rRNA (). Considering this, it is tempting to speculate that SURF6 is involved in the cleavage of 36S rRNA along the ITS2 spacer (). Northern blots also showed that an excess of SURF6 causes an accumulation of 45S pre-rRNA () that is formed downstream from the 47S pre-rRNA precursor (). This may result either from retardation of 45S pre-rRNA processing or from activation of the 47S pre-rRNA synthesis. The later assumption sounds more probable because the number of amplicons corresponding to the shortly lived 5′ETS fragments determined by qRT-PCR points to a 1.5-fold increase of the 47S pre-rRNA copies in +Dox fibroblasts as compared with –Dox cells (). The increase of 18S, 5.8S and 28S rRNA amplicons also detected by qRT-PCR () is consistent with the Northern blot data, evidencing in favor that SURF6 is involved in the maturation of both subunits at the point of rRNA processing. (). In addition, the accumulation of these amplicons may arise from an increasing level of the cytoplasmic rRNAs, which were not specifically identified on the blots.
Overall, our work concludes that the nucleolar protein SURF6 is involved in the maturation of both ribosomal subunits at the point of rRNA processing. SURF6 can stimulate proliferation of sub-confluent NIH/3T3-174 fibroblasts by decreasing their population doubling-time, shortening the cell division cycle and by facilitating G1/S transition, and thereby serves as a positive regulator of proliferation in non-malignant mammalian cells. Our results support the idea that SURF6 could be a putative oncoprotein Citation31 and an appropriate candidate for anti-cancer therapy.Citation44
Material and methods
Cells and DNA plasmid constructs
Mouse embryo NIH/3T3 fibroblasts were purchased from the Russian Collection of Cell Cultures (Institute of Cytology, Russian Academy of Sciences, St. Petersburg, Russia). Cells were grown in Dulbecco's modified Eagle's medium (Sigma-Aldrich, USA) supplemented with 10% fetal calf serum (FCS; HyClone, USA), 4.5 g/L glucose, 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, penicillin, and streptomycin (100 units/ml each) at 37° C and the atmosphere of 5% CO2/95% air.
Stably transfected NIH/3T3-174 fibroblasts were established by the use of two plasmid constructs, which are components of the Tet-On® gene expression system (Clontech Laboratories, Inc., USA). The first (auxiliary) plasmid pcDNA3.1(-)rtTA encodes the tetracycline-controlled transactivator and the neomycin (G418) resistance gene. It was created by cloning the reverse transcriptional transactivator (rtTA) cDNA from the pUHrT62-1 plasmidCitation45 into the EcoRI/BamHI cloning sites of the pcDNA3.1(-) vector (Invitrogen, USA). The second (executive) plasmid, named as pBI-mSURF6, was engineered using a commercial vector pBI-EGFP (Clontech Laboratories, Inc.). The NheI/NheI 1.9 kb cDNA fragment, corresponding to the mouse SURF6 coding region, was cut off from the pBS-Surf6 cloneCitation25 and re-cloned to the NheI cloning sites of the pBI-EGFP vector. The resulted pBI-mSURF6 plasmid contains a bi-directorial TRE (tetracycline-responsible element) activated by the rtTA expression product flanked by two minimal bi-directorial CMV promoters which govern co-expression of SURF6 and EGFP ().
Transfections were performed with the Lipofectamine2000 reagent (Invitrogen, USA). NIH/3T3 fibroblasts were transfected with the pcDNA3.1(-)rtTA plasmid, and the transfectants were selected by one-month culturing in the presence of 450 µg/ml G418. Selected clones were co-transfected with the plasmid pBI-mSURF6 (to generate NIH/3T3-174 fibroblasts) or with the pBI-EGFP vector (to generate NIH/3T3-EGFP cells capable to express EGFP alone) mixed in the equal amounts with a linear DNA fragment containing the puromycin-resistance gene. The DNA fragment of the puromycin-resistance gene was excised from the plasmid pLoxPuro with BamHI.Citation46 The transfectants were cultured in complete DMEM supplemented with 200 µg/ml G418 and 1 µg/ml puromycin for another month. Clones resistant to the second selection were transferred to wells of a 96-well plate and allowed growing. Most healthy clones were named as NIH/3T3-174 and NIH/3T3-EGFP fibroblasts and used in the study.
The stably transfected cells were cultured in DMEM medium containing 10% Tet-On approved fetal bovine serum (FBS, Clontech Laboratories, Inc.), 4.5 g/l glucose, 2 mM L-glutamine, 1.5 g/l sodium bicarbonate, penicillin and streptomycin (100 units/ml each), 200 µg/ml G418 and 0.5 µg/ml puromycin at 37° C and in the atmosphere of 5% CO2/95%. Cells were plated with initial density of 3–5 × 103 cell/сm2 unless indicated otherwise. Doxycycline chloride (Sigma-Aldrich, USA) was dissolved in DMEM to the final concentrations 10, 100 or 1000 ng/ml and added to sub-confluent cells cultured in complete growth medium for 3–24 hours prior the induction. In control experiments (-Dox, non-induced cells), the equal volume of DMEM alone was added. The initial cell density was counted with an automated cell counter Cedex XS Analyzer (Roche, USA).
Immunoblotting and immunofluorescence
For immunoblots, 5 × 106 cells were lysed in 500 µl buffer containing 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 10% glycerol, 0.5% Triton X-100 and a cocktail of protease inhibitors (Sigma-Aldrich) on ice. The total protein concentration in samples was determined with the Lowry-Peterson assay using a Protein Assay Kit (Sigma-Aldrich). Fifty µg of proteins in Laemmli buffer (60 mМ Tris-Cl, рН 6.8, 10% glycerol, 2% SDS, 100 mM β-mercaptoethanol, 0.01% bromophenol blue) was loaded in every lane of 12% SDS-PAGE. Blotted nitrocellulose membranes were incubated with the rabbit serum specifically recognizing mouse SURF6Citation26 or with a mouse monoclonal antibody to beta-tubulin (Sigma-Aldrich) served as a loading control, and then with relevant horseradish peroxidase–conjugated antibodies diluted in TBST buffer (20 mМ Трис, рН 7.6, 150 mМ NaCl, 0.05% Tween-20) supplemented with 5% skim milk. The proteins were recovered using an ECL+Plus Kit (Amersham Pharmacia Biotech Inc., USA). Quantification of the immunoblot signals was performed with TotalLab Quantification Software (TotalLab Limited, UK).
For immunofluorescence, NIH/3T3-174 fibroblasts grown on coverslips were fixed with 2% paraformaldehyde in phosphate buffered saline (PBS, 140 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, and 8.1 mM Na2HPO4, рН 7.2) for 20 min at room temperature. Then, they were treated with 0.5% Triton X-100 in PBS for 10 min at room temperature, and labeled with the anti-SURF6 serumCitation26 followed by AlexaFluor 488 goat anti-rabbit IgG (H+L) (Molecular Probes Inc., USA, cat.A-11034). Cells were counterstained with a DNA-binding dye DAPI (1 μg/ml in PBS, 10 min), mounted in Vectashield® (Vector Laboratories, USA), and examined with a DuoScanMeta LSM510 confocal laser scanning microscope (Carl Zeiss, Germany) equipped with a Plan-Apochromat 63 × /1.40 (numerical aperture) oil Ph3 objective.
Analysis of cell growth with the xCELLigence System
The xCELLigence Real-Time Cell Analysis (RTCA)-DP system (ACEA Biosciences, CAL, USA) was used to monitor cell growth in real time according to.Citation47 The technology measures impedance-based signals, which occur through cell contacts with microelectrodes, and the signal values are interpreted as normalized cell indexes. NIH3T3-174 cells were seeded in duplicate in an E-16 plate (ACEA Biosciences), cultured under standard conditions for ∼24 hours before adding 10–1000 ng/ml doxycycline in DMEM or DMEM alone (the –Dox controls). Impedimetric signals were recorded automatically every 15 min for ∼3 days. The cell growth rate was determined by calculating the slope of the curves between the time-point of doxycycline administration (∼24 hrs) and the time-point corresponding to the end of an experiment (∼70 hrs). Experiments were repeated several times, and the results are expressed as the mean ± SEM (Standard Error Mean).
MTT assay
NIH/3T3-174 fibroblasts were seeded in 24-well plates in complete growth medium for 3 hours to be recovered. Cells were rinsed three times with PBS and incubated in serum-free medium for 24 hours, and afterwards 100 ng/ml doxycycline in DMEM (+Dox cells) or the equal volume of DMEM (-Dox cells) were added as described.Citation48 The MTT assay was executed as recommended by the manufacture (Life Technologies, France) shortly after doxycycline administration (“0” time-point) and also 24 and 48 hours later. Absorbance was read at 540 nm with a Thermo Scientific Multiskan EX ELISA reader (Thermo Fisher Scientific, USA). Experiments were repeated in triplicate.
Calculation of the population doubling time
Cells were seeded in 6 cm Petri dishes and after culturing for 3–4 hours, 100 ng/ml doxycycline in DMEM or DMEM alone (-Dox control) were added (time-point “0”, T0). Twenty five random fields of view were photographed at the “0” point and then every 24 hours (Tt) under a microscope Axiovert 200 (Carl Zeiss, Germany) coupled with a 8-bit digital CCD camera CoolSnapcf (Photometrics, USA) and a Plan-Neofluar 10 × /NA0.4 Ph lens. The average cell population doubling time (td) was calculated with the formula (No Equation1(1)
(1) ):
(1)
(1) where t is an interval (in hours) between T0 and Tt, and N0 and Nt are the average cell number per microscopic field of view at the same time-points.Citation49 Experiments were repeated five times.
Flow cytometry
Cells were fixed with 70% ethanol for 15 min at 4о С and stained with 50 µg/ml propidium iodide (Sigma-Aldrich) in PBS containing 0.5 mg/ml bovine pancreatic RNAase A (Sigma-Aldrich) for 1 h at 37о С. Above 1 × 104 cells were analyzed in each sample using a flow cytometer Epics ”Elite” (Beckman Coulter Inc., USA) equipped with an argon laser Cyonics (Uniphase, USA) and Multigraph software (Coulter, USA). Experiments were repeated four times.
Timing of cell cycle and the cell cycle phases
The duration of the cell division cycle (T, hours) was calculated according to Citation49 with the equation (No Equation2(2)
(2) ):
(2)
(2) where td – the average cell population doubling time, and y – the average percentage of G0 cells (G0 index).
To determine the G0 index, cells were seeded on coverslips, cultured overnight and incubated with 10 mМ Br-dU (-Dox control cells) or 10 mМ Br-dU + 100 ng/ml doxycycline (+Dox cells) for 24 hours, a period that exceeds the division cycle in 3T3 fibroblasts.Citation50 The cells were fixed with 4 % paraformaldehyde in PBS for 20 min, permeabilized with 0.5% Triton Х-100 in PBS for 10 min, and treated with 2М HCl for 40 min at room temperature. Cells were intensely washed in PBS, immersed in a mouse anti-Br-dU monoclonal antibody (Roche, USA) for 45 min and then to Texas Red-conjugated antibodies to mouse IgG (Jackson ImmunoResearch Labs, Inc., USA) for 45 min at 37° С. Cells were counterstained with DAPI (4′,6-Diamidino-2-phenylindole dihydrochloride) and mounted in Vectashield® (Vector Laboratories, USA). Samples were examined under an Axiovert200 epifluorescence microscope equipped with a Plan-NeoFluar × 40/NA 0.75 Ph lens. Br-dU-negative cells were considered as G0, or steady-state, cells. Above 2,000 cells were analyzed at each experimental and control time-points. G0 index was defined as the ratio between the number of G0 cells to the total number of cells. The sum duration of G2 phase and mitosis (tG2+M, hours) was calculated with the formula (No Equation3(3)
(3) )Citation51:
(3)
(3) where (G2+M)/N – the percentage of cells in G2 phase and mitosis, y' – the percentage of proliferating cells equals to the total number of cells minus G0 cells. The percentage of G2 and mitotic cells, i.e., G2/M cells, was determined by flow cytometry.
The length of S phase (ts, hours) was calculated with the equation (No Equation4(4)
(4) )Citation51:
(4)
(4) where T – the duration of cell cycle in hours, S/N – the percentage of S phase cells, (G2+M)/N – the percentage of G2+mitotic cells, y' – the percentage of proliferating cells. The percentage of S phase cells was determined by flow cytometry.
The length of G1 phase (tG1, hours) was calculated with the formula (No Equation5(5)
(5) )Citation51:
(5)
(5) where T is the durations of cell division cycle, tS is the duration of S phase, and tG2+M is the sum duration of G2 phase and mitosis determined according to the formula (Equation3
(3)
(3) ).
Mitotic index, or the ratio between the number of mitotic cells and the total number of cells, was counted using DAPI-stained cells, a microscope Axiovert 200 and a Plan-NeoFluar × 40/NA 0.75 Ph lens. Above 2,000 cells were analyzed at each control and experimental point.
Northern blot analysis
Approximately 1 × 106 cells were lysed with 1 ml TRIzol® Reagent (Life Technologies, USA) and RNA was isolated as recommended by the manufacture. In all samples, the ratio А260/А280 was within the range of 1.9 to 2.0 thus indicating the absence of protein impurities. RNA was quantified with a nanodrop spectrophotometer (Thermo-Scientific, MA, USA). An equal amount of RNA was loaded per well and processed for Northern blotting as described previously.Citation52 Five micrograms of total RNAs were fractionated on standard 1% agarose/6% formaldehyde gels to analyse RNA species. Fifty pmol of a probe was mixed with 50 pmol of [γ-32P]ATP and T4 polynucleotide kinase (Promega, France) and incubated for 30 min at 37° C. For each experimental condition, the radioactivity contained in each band was quantified with a PhosphorImager (Typhoon, GE Healthcare, USA). Two independent experiments were performed. The values obtained in +Dox NIH/3T3-174 fibroblasts (100 ng/ml doxycycline, 24 hrs of the treatment) were averaged on the basis of four Northern blot data and expressed as the mean values normalized to the control mean values obtained in –Dox fibroblasts. Statistical analysis (one sample t-test) from results obtained in independent experiments was performed with RStudio software (https://www.rstudio.com/products/rstudio/).
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was isolated from –Dox and +Dox fibroblast after 24 hours of 100 ng/ml doxycycline administration and used to determine expression of rRNA species as described in.Citation53 The primers and FAM-conjugated probes targeting the short-lived 5′ETS and the 18S, 5.8S and 28S regions of the mouse 47S pre-rRNA (GenBank ID: X82564), Surf6 mRNA (GenBank ID: NM_009298) and beta-actin mRNA (GenBank ID: NM_007393) were designed with AlleleID 7.7 software (Premier Biosoft, USA) and synthesized by DNA-Synthesis LLC (Moscow, Russia) (). Beta-actin mRNA served as the internal reference control. The master mix, qPCRmix-HS (Evrogen, Moscow, Russia), and 2 μl of cDNA were used in a PCR reaction. qRT-PCR was performed five times for each target amplicon (i.e., 5′ETS, 18S, 5.8S and 28S rRNA, Surf6 mRNA and beta-actin mRNA) using a MiniOpticon RT-PCR system (BioRad, Hercules, USA) with the following parameters: 5 min cDNA denaturation at 95o C and 40 PCR cycles each for 15 s at 95o C, 20 s at 55o C and 20 s at 72o C. Melting curve analysis and gel electrophoresis were used to control the specificity and the quality of PCR. The data were analyzed with Bio-Rad CFX software (BioRad). Fold-induction values (N) were calculated for each target amplicon as described inCitation54 with the formula:
Table 3. Primers and FAM-conjugated probes used in qRT-PCR analysis of the amplified DNA products (amplicons) corresponding to the mouse 47S pre-rRNA transcripts, 18S, 5.8S and 28S rRNAs, beta-actin and SURF6 mRNAs.
Herein, ΔΔCt represents the difference between ΔCt+dox and ΔCt-dox values for each given amplicon, ΔCt is the difference between Cttarget and Ctreference, and Ct is the mean threshold cycle for each target and the reference amplicons. The results of five qRT-PCR independent experiments were averaged.
Statistical analysis
Results are expressed as mean ± standard error of the mean (SEM). Control and experimental values were compared with the Student t-test assuming unequal variances. Differences are considered significant at a p value less than 0.05. Microsoft Excel 2007 software (Microsoft, Redmond, WA, USA) was used for statistical analysis of data.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The study was supported by the Russian Foundation for Basic Research (grant 16-04-01199) and partly by a collaborative program between CNRS (France) and RAS (Russia).
References
- Mullineux S-T, Lafontaine DLJ. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie. 2012;94:1521-32. doi:10.1016/j.biochi.2012.02.001. PMID:22342225
- Coleman AW. Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends Genet. 2015;31:157-63. doi:10.1016/j.tig.2015.01.002. PMID:25648500
- Montanaro L, Treré D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol. 2008;173:301-10. doi:10.2353/ajpath.2008.070752. PMID:18583314
- Brighenti E, Treré D, Derenzini M. Targeted cancer therapy with ribosome biogenesis inhibitors: a real possibility? Oncotarget. 2015;6:38617-27. doi:10.18632/oncotarget.5775. PMID:26415219
- Takada H, Kurisaki A. Emerging roles of nucleolar and ribosomal proteins in cancer, development, and aging. Cell Mol Life Sci. 2015;72:4015-25. doi:10.1007/s00018-015-1984-1. PMID:26206377
- Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD. Targeting the nucleolus for cancer intervention. Biochim Biophys Acta. 2014;1842:802-16. doi:10.1016/j.bbadis.2013.12.009. PMID:24389329
- Tsai RYL, Pederson T. Connecting the nucleolus to the cell cycle and human disease. FASEB J. 2014;28:3290-6. doi:10.1096/fj.14-254680. PMID:24790035
- Woods SJ, Hannan KM, Pearson RB, Hannan RD. The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochim Biophys Acta. 2015;1849:821-9. doi:10.1016/j.bbagrm.2014.10.007. PMID:25464032
- Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin Cancer Biol. 2016;37-38: 36-50. doi:10.1016/j.semcancer.2015.12.004. PMID:26721423
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673-83. doi:10.1016/j.bbamcr.2009.10.009. PMID:19879902
- Grummt I. The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122:487-97. doi:10.1007/s00412-013-0430-0. PMID:24022641
- Li P, Wang D, Li H, Yu Z, Chen X, Fang J. Identification of nucleolus-localized PTEN and its function in regulating ribosome biogenesis. Mol Biol Rep. 2014;41:6383-90. doi:10.1007/s11033-014-3518-6. PMID:24969487
- Durut N, Sáez-Vásquez J. Nucleolin: dual roles in chromatin transcription. Gene. 2015;556:7-12. doi:10.1016/j.gene.2014.09.023. PMID:25225127
- Rohrmoser M, Hölzel M, Grimm T, Malamoussi A, Harasim T, Orban M, Pfisterer I, Gruber-Eber A, Kremmer E, Eick D. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol. 2007;27:3682-94. doi:10.1128/MCB.00172-07. PMID:17353269
- Ahn CS, Cho HK, Lee D-H, Sim H-J, Kim S-G, Pai H-S. Functional characterization of the ribosome biogenesis factors PES, BOP1, and WDR12 (PeBoW), and mechanisms of defective cell growth and proliferation caused by PeBoW deficiency in Arabidopsis. J Exp Bot. 2016;67:5217-32. doi:10.1093/jxb/erw288. PMID:27440937
- Zentner GE, Hurd EA, Schnetz MP, Handoko L, Wang C, Wang Z, Wei C, Tesar PJ, Hatzoglou M, Martin DM, et al. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum Mol Genet. 2010;19:3491-501. doi:10.1093/hmg/ddq265. PMID:20591827
- Castle CD, Cassimere EK, Lee J, Denicourt C. Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol Cell Biol. 2010;30:4404-14. doi:10.1128/MCB.00358-10. PMID:20647540
- Zhang Y, Forys JT, Miceli AP, Gwinn AS, Weber JD. Identification of DHX33 as a mediator of rRNA synthesis and cell growth. Mol Cell Biol. 2011;31:4676-91. doi:10.1128/MCB.05832-11. PMID:21930779
- Ardestani PM, Liang F. Sub-cellular localization, expression and functions of Sirt6 during the cell cycle in HeLa cells. Nucleus. 2012;3:442-51. doi:10.4161/nucl.21134. PMID:22743824
- Wang J, Leung JW, Gong Z, Feng L, Shi X, Chen J. PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. J Biol Chem. 2013;288:3174-83. doi:10.1074/jbc.M112.414839. PMID:23229552
- Zhang H, Ma X, Shi T, Song Q, Zhao H, Ma D. NSA2, a novel nucleolus protein regulates cell proliferation and cell cycle. Biochem Biophys Res Commun. 2010;391:651-8. doi:10.1016/j.bbrc.2009.11.114. PMID:19932687
- Naora H, Takai I, Adachi M, Naora H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741-53. doi:10.1083/jcb.141.3.741. PMID:9566973
- Kim J-H, You K-R, Kim IH, Cho B-H, Kim C-Y, Kim D-G. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology. 2004;39:129-38. doi:10.1002/hep.20017. PMID:14752831
- Polzikov M, Zatsepina O, Magoulas C. Identification of an evolutionary conserved SURF-6 domain in a family of nucleolar proteins extending from human to yeast. Biochem Biophys Res Commun. 2005;327:143-9. doi:10.1016/j.bbrc.2004.11.148. PMID:15629442
- Magoulas C, Fried M. The Surf-6 gene of the mouse surfeit locus encodes a novel nucleolar protein. DNA Cell Biol. 1996;15:305-16. doi:10.1089/dna.1996.15.305. PMID:8639267
- Magoulas C, Zatsepina O V, Jordan PW, Jordan EG, Fried M. The SURF-6 protein is a component of the nucleolar matrix and has a high binding capacity for nucleic acids in vitro. Eur J Cell Biol. 1998;75:174-83. doi:10.1016/S0171-9335(98)80059-9. PMID:9548374
- Ringwald M, Wu C, Su AI. BioGPS and GXD: mouse gene expression data-the benefits and challenges of data integration. Mamm Genome. 2012;23:550-8. doi:10.1007/s00335-012-9408-0. PMID:22847375
- Moraleva AA, Malysheva M V, Magoulas C, Polzikov MA, Zatsepina O V. Early expression of nucleolar SURF-6 protein in mouse spleen lymphocytes activated for proliferation in vitro. Bull Exp Biol Med. 2009;147:578-82. doi:10.1007/s10517-009-0578-z. PMID:19907743
- Oeffinger M, Fatica A, Rout MP, Tollervey D. Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis. Nucleic Acids Res. 2007;35:1354-66. doi:10.1093/nar/gkl824. PMID:17272295
- Yamada H, Horigome C, Okada T, Shirai C, Mizuta K. Yeast Rrp14p is a nucleolar protein involved in both ribosome biogenesis and cell polarity. RNA. 2007;13:1977-87. doi:10.1261/rna.553807 doi:10.1261/rna.553807. PMID:17804645
- Wan D, Gong Y, Qin W, Zhang P, Li J, Wei L, Zhou X, Li H, Qiu X, Zhong F, et al. Large-scale cDNA transfection screening for genes related to cancer development and progression. Proc Natl Acad Sci U S A. 2004;101:15724-9. doi:10.1073/pnas.0404089101. PMID:15498874
- Polzikov M, Magoulas C, Zatsepina O. The nucleolar protein SURF-6 is essential for viability in mouse NIH/3T3 cells. Mol Biol Rep. 2007;34:155-60. doi:10.1007/s11033-006-9028-4. PMID:17086444
- Lafarga M, Lerga A, Andres MA, Polanco JI, Calle E, Berciano MT. Apoptosis induced by methylazoxymethanol in developing rat cerebellum: organization of the cell nucleus and its relationship to DNA and rRNA degradation. Cell Tissue Res. 1997;289:25-38. doi:10.1007/s004410050849. PMID:9182598
- Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ. Is green fluorescent protein toxic to the living cells? Biochem Biophys Res Commun. 1999;260:712-7. doi:10.1006/bbrc.1999.0954. PMID:10403831
- Xie J, Nair A, Hermiston TW. A comparative study examining the cytotoxicity of inducible gene expression system ligands in different cell types. Toxicol In Vitro. 2008;22:261-6. doi:10.1016/j.tiv.2007.08.019. PMID:17928190
- Li Z, Hann SR. Nucleophosmin is essential for c-Myc nucleolar localization and c-Myc-mediated rDNA transcription. Oncogene. 2013;32:1988-94. doi:10.1038/onc.2012.227. PMID:22665062
- Tsai RYL, McKay RDG. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991-3003. doi:10.1101/gad.55671. PMID:12464630
- Romanova L, Grand A, Zhang L, Rayner S, Katoku-Kikyo N, Kellner S, Kikyo N. Critical role of nucleostemin in pre-rRNA processing. J Biol Chem. 2009;284:4968-77. doi:10.1074/jbc.M804594200. PMID:19106111
- Kondo T, Minamino N, Nagamura-Inoue T, Matsumoto M, Taniguchi T, Tanaka N. Identification and characterization of nucleophosmin/B23/numatrin which binds the anti-oncogenic transcription factor IRF-1 and manifests oncogenic activity. Oncogene. 1997;15:1275-81. doi:10.1038/sj.onc.1201286. PMID:9315094
- Yu W, Qiu Z, Gao N, Wang L, Cui H, Qian Y, Jiang L, Luo J, Yi Z, Lu H, et al. PAK1IP1, a ribosomal stress-induced nucleolar protein, regulates cell proliferation via the p53-MDM2 loop. Nucleic Acids Res. 2011;39:2234-48. doi:10.1093/nar/gkq1117. PMID:21097889
- Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766-9. doi:10.1126/science.7792603. PMID:7792603
- Kordyukova MY, Polzikov MA, Shishova K V, Zatsepina O V. Functional significance of the human nucleolar protein SURF6, the key member of the SURF6 protein family in eukaryotes. Dokl Biochem Biophys. 2014;455:65-7. doi:10.1134/S1607672914020069. PMID:24795102
- Hannan KM, Kennedy BK, Cavanaugh AH, Hannan RD, Hirschler-Laszkiewicz I, Jefferson LS, Rothblum LI. RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene. 2000;19:3487-97. doi:10.1038/sj.onc.1203690. PMID:10918607
- Chen K, Ma H, Li L, Zang R, Wang C, Song F, Shi T, Yu D, Yang M, Xue W, et al. Genome-wide association study identifies new susceptibility loci for epithelial ovarian cancer in Han Chinese women. Nat Commun. 2014;5:4682. doi:10.1038/ncomms5682. PMID:25134534
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci U S A. 2000;97:7963-8. doi:10.1073/pnas.130192197. PMID:10859354
- Arakawa H, Lodygin D, Buerstedde J-M. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 2001;1:7. doi:10.1186/1472-6750-1-7. PMID:11591226
- Vouyovitch CM, Perry JK, Liu DX, Bezin L, Vilain E, Diaz J-J, Lobie PE, Mertani HC. WNT4 mediates the autocrine effects of growth hormone in mammary carcinoma cells. Endocr Relat Cancer. 2016;23:571-85. doi:10.1530/ERC-15-0528. PMID:27323961
- Chiesa J, Ferrer C, Arnould C, Vouyovitch CM, Diaz J-J, Gonzalez S, Mares P, Morel G, Wu Z-S, Zhu T, et al. Autocrine proliferative effects of hGH are maintained in primary cultures of human mammary carcinoma cells. J Clin Endocrinol Metab. 2011;96:E1418-26. doi:10.1210/jc.2011-0473. PMID:21733992
- Uzbekov RE. Analysis of the cell cycle and a method employing synchronized cells for study of protein expression at various stages of the cell cycle. Biochemistry (Moscow). 2004;69:485-96. doi:10.1023/B:BIRY.0000029845.11184.30
- Brooks RF, Riddle PN. The 3T3 cell cycle at low proliferation rates. J Cell Sci. 1988;90 (Pt 4):601-12. PMID:3253297
- Watanabe I, Okada S. Effects of temperature on growth rate of cultured mammalian cells (L5178Y). J Cell Biol. 1967;32:309-23. doi:10.1083/jcb.32.2.309. PMID:10976224
- Couté Y, Kindbeiter K, Belin S, Dieckmann R, Duret L, Bezin L, Sanchez J-C, Diaz J-J. ISG20L2, a novel vertebrate nucleolar exoribonuclease involved in ribosome biogenesis. Mol Cell Proteomics. 2008;7:546-59. doi:10.1074/mcp.M700510-MCP200. PMID:18065403
- Polzikov M, Yakovenko S, Voznesenskaya J, Troshina M, Zatsepina O. Overexpression of ribosomal RNA in cumulus cells of patients with polycystic ovary syndrome. J Assist Reprod Genet. 2012;29:1141-5. doi:10.1007/s10815-012-9827-6. PMID:22772454
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101-8. doi:10.1038/nprot.2008.73. PMID:18546601
