ABSTRACT
Programmed cell death 4 (Pdcd4) is frequently suppressed in tumors of various origins and its suppression correlates with tumor progression. Pdcd4 inhibits cap-dependent translation from mRNAs with highly structured 5′-regions through interaction with the eukaryotic translation initiation factor 4A (eIF4A) helicase and a target transcript. Decrease in Pdcd4 protein is believed to provide a relief of otherwise suppressed eIF4A-dependent translation of proteins facilitating tumor progression. However, it remains unknown if lowered Pdcd4 levels in cells suffices to cause a relief in translation inhibition through appearance of the Pdcd4-free translation-competent eIF4A protein, or more complex and selective mechanisms are involved. Here we showed that eIF4A1, the eIF4A isoform involved in translation, significantly over-represents Pdcd4 both in cancerous and normal cells. This observation excludes the possibility that cytoplasmic Pdcd4 can efficiently exert its translation suppression function owing to excess of eIF4A, with Pdcd4-free eIF4A being in excess over Pdcd4-bound translation-incompetent eIF4A, thus leaving translation from Pdcd4 mRNA targets unaffected. This contradiction is resumed in the proposed model, which supposes initial complexing between Pdcd4 and its target mRNAs in the nucleus, with subsequent transport of translation-incompetent, Pdcd4-bound target mRNAs into the cytoplasm. Noteworthy, loss of nuclear Pdcd4 in cancer cells was reported to correlate with tumor progression, which supports the proposed model of Pdcd4 functioning.
Programmed cell death 4 (Pdcd4) is a tumor suppressor inhibiting cap-dependent translation from mRNAs with highly structured 5′-regions.Citation1,Citation2 Pdcd4-mediated translational suppression relies on its interaction with eIF4A helicase (namely, eIF4A1 and eIF4A2)Citation2 resulting in inhibition of eukaryotic translation initiation factor 4A (eIF4A) helicase activity and precluding from eIF4A:eIF4G complex formation, and, consequently, eIF4F translation initiation factor assembly.Citation3-6 Pdcd4 is down-regulated in tumor cells by several mechanisms including ubiquitin-dependent protein degradation,Citation6,Citation7 microRNA-dependent suppression of translation from and degradation of Pdcd4 mRNA (for example,Citation9-11), suppression of Pdcd4 gene transcription.Citation12 Lowered Pdcd4 protein level is believed to provide a relief to otherwise suppressed production of proteins facilitating tumor progression which translation depends on eIF4A,Citation13-15 resulting in increased cell motility and invasion, enhanced proliferative potential, suppression of apoptotic pathways, drug resistance.Citation1,Citation2
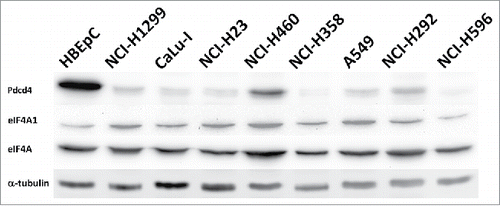
Along with eIF4A binding, Pdcd4 also interacts with mRNA molecules, forming a tri-partite (Pdcd4:mRNA:eIF4A) complex which is non-permissive for mRNA translation.Citation16-18 However, it remains unknown if lowered Pdcd4 levels in cells per se suffices to cause a relief in translation inhibition through an increase in a pool of Pdcd4-free translation-competent eIF4A protein, or more complex and selective mechanisms are involved.
To test the hypothesis that decrease in Pdcd4 level per se is sufficient to relief eIF4A-dependent translation, we assessed relative abundances of Pdcd4 and eIF4A1, one of the two eIF4A isoforms involved in translation initiation, in cell. When precipitated from the total lysate of NCI-H1299 lung cancer cells stably producing enhanced green fluorescent protein (EGFP)-tagged Pdcd4 protein (NCI-H1299[E-Pdcd4] cells), eIF4A1 was the major EGFP-Pdcd4 binding partner as determined by matrix-assisted laser desorption/ionization-time of flight mass-spectrometry (MALDI-TOF MS) (rank #1 in the spectrum analysis report, with 40.6% sequence coverage; ). Pdcd4 is known to bind eIF4A in solution with 1:2 binding stoichiometry between Pdcd4 and eIF4A.Citation5 Comparable intensities of eIF4A and EGFP-Pdcd4 protein bands () and approximately 2-fold difference in EGFP-Pdcd4 and eIF4A molecular weights fit 1:2 stoichiometry between Pdcd4 and eIF4A in the immunoprecipitate and suggest that all the EGFP-Pdcd4 protein in the lysate is complexed with eIF4A, with no eIF4A-free EGFP-Pdcd4 protein being present which would result in over-representation of EGFP-Pdcd4 protein over eIF4A protein in the immuneprecipitate. However, nearly complete depletion of EGFP-Pdcd4 protein from the lysate resulted in a subtle change in eIF4A1 protein amount in the flow-through fraction compared to the initial lysate (). As apparently all the EGFP-Pdcd4 protein in the lysate is complexed with eIF4A as reasoned above, the eIF4A1 protein in the flow-through represents an excess of the eIF4A1 protein over EGFP-Pdcd4, with no free EGFP-Pdcd4 protein in the lysate which could bind it. EGFP-Pdcd4 protein level in NCI-H1299[E-Pdcd4] cells exceeds that of endogenous Pdcd4 protein (see , lane 1), with gross over-representation of eIF4A1 over EGFP-Pdcd4. Therefore, tumor cells have an excess of eIF4A1 over Pdcd4, and even if all the Pdcd4 protein would bind eIF4A1, an abundant pool of Pdcd4-free and thus competent for translation initiation eIF4A1 protein would remain in tumor cells. This can be caused by down-regulation of Pdcd4 (reviewed inCitation1,Citation2,Citation19), up-regulation of eIF4A,Citation20 or both. While Pdcd4 is suppressed to various extents in NCI-H1299 and other lung cancer cells ( and Ref.Citation12,Citation21), total levels of eIF4A protein and eIF4A1 protein levels in particular, are not significantly altered in cancerous cells compared to primary human bronchial epithelial cells (HBEpC; ). Indeed, when relative abundances of eIF4A1 and Pdcd4 proteins were assessed in similar way in non-cancerous HBEpC cells, a gross over-representation of eIF4A1 was also observed (). Thus, the simple mechanistical model of Pdcd4 action through exhaustion of translation-competent eIF4A proteins is void.
Figure 1. (A) eIF4A1 is the major Pdcd4 binding partner in NCI-H1299 cells. NCI-H1299[E-Pdcd4] clonal derivative of NCI-H1299 cells (ATCC #CRL-5803) was obtained by transfection of the cells with pEGFP-Pdcd4 plasmidCitation21 with subsequent selection of clones stably producing EGFP-Pdcd4 protein. Lysates of NCI-H1299[E-Pdcd4] (lane 1) or parental NCI-H1299 cells (lane 2) were prepared in the buffer (25 mM Tris-HCl, pH 7.4; 250 mM NaCl; 5 mM EDTA; 1% NP-40; 1x Protease inhibitor cocktail (Sigma, St. Louis, MO, USA)) and incubated with anti-EGFP monoclonal antibody-coated beads (Proteinsynthesis, Moscow, Russia). After washing with phosphate buffered saline, captured proteins were eluted in SDS-PAAG loading buffer by boiling and separated in 10% SDS-PAAG. The gel was stained with Bio-Safe Coomassie stain (Bio-Rad, Hercules, CA, USA). Approximately 50 kDa protein band (marked by an arrow) co-purified with EGFP-Pdcd4 protein (marked by an asterisk) from NCI-H1299[E-Pdcd4] cell extract was excised from the gel and identified as human eIF4A1 protein based on results of MALDI-TOF mass-spectrometry. MW – protein molecular weight marker, with protein molecular weights indicated in kDa on the right. (B) Evaluation of relative Pdcd4 and eIF4A1 protein abundances in cancerous NCI-H1299 cells. Lysate of NCI-H1299[E-Pdcd4] cells was incubated with anti-EGFP monoclonal antibody-coated beads. After incubation, bead washing and protein elution from beads as described in (A), proportional amounts of initial lysates (lanes 1), eluted proteins (lanes 2) and flow-through (lanes 3) were analyzed by Western blotting with anti-Pdcd422 or anti-eIF4A1 (Cell Signaling, Boston, MA, USA) rabbit antibodies. EGFP-Pdcd4 and endogenous Pdcd4 protein bands are marked by an asterisk and by an arrow, respectively. (C) Evaluation of relative Pdcd4 and eIF4A1 protein abundances in non-cancerous HBEpC cells. Lysate of HBEpC cells (EACCC #502-05a) infected with recombinant replication-deficient adenovirus for EGFP-Pdcd4 protein production (constructed using Ad-Easy system (Stratagene, La Jolla, CA, USA) with derived from pEGFP-Pdcd4 plasmid expression module consisting from CMV promoter-driven EGFP-Pdcd4 cDNA with SV40 virus transcription termination and poyadenylation signal) was incubated with anti-EGFP monoclonal antibody-coated beads. After incubation, bead washing and protein elution from beads as described in (A), proportional amounts of initial lysates (lanes 1), eluted proteins (lanes 2) and flow-through (lanes 3) were analyzed by Western blotting with anti-Pdcd421 or anti-eIF4A1 (Cell Signaling, Boston, MA, USA; Cat. #2490) rabbit antibodies. EGFP-Pdcd4 protein band is marked by an asterisk.
![Figure 1. (A) eIF4A1 is the major Pdcd4 binding partner in NCI-H1299 cells. NCI-H1299[E-Pdcd4] clonal derivative of NCI-H1299 cells (ATCC #CRL-5803) was obtained by transfection of the cells with pEGFP-Pdcd4 plasmidCitation21 with subsequent selection of clones stably producing EGFP-Pdcd4 protein. Lysates of NCI-H1299[E-Pdcd4] (lane 1) or parental NCI-H1299 cells (lane 2) were prepared in the buffer (25 mM Tris-HCl, pH 7.4; 250 mM NaCl; 5 mM EDTA; 1% NP-40; 1x Protease inhibitor cocktail (Sigma, St. Louis, MO, USA)) and incubated with anti-EGFP monoclonal antibody-coated beads (Proteinsynthesis, Moscow, Russia). After washing with phosphate buffered saline, captured proteins were eluted in SDS-PAAG loading buffer by boiling and separated in 10% SDS-PAAG. The gel was stained with Bio-Safe Coomassie stain (Bio-Rad, Hercules, CA, USA). Approximately 50 kDa protein band (marked by an arrow) co-purified with EGFP-Pdcd4 protein (marked by an asterisk) from NCI-H1299[E-Pdcd4] cell extract was excised from the gel and identified as human eIF4A1 protein based on results of MALDI-TOF mass-spectrometry. MW – protein molecular weight marker, with protein molecular weights indicated in kDa on the right. (B) Evaluation of relative Pdcd4 and eIF4A1 protein abundances in cancerous NCI-H1299 cells. Lysate of NCI-H1299[E-Pdcd4] cells was incubated with anti-EGFP monoclonal antibody-coated beads. After incubation, bead washing and protein elution from beads as described in (A), proportional amounts of initial lysates (lanes 1), eluted proteins (lanes 2) and flow-through (lanes 3) were analyzed by Western blotting with anti-Pdcd422 or anti-eIF4A1 (Cell Signaling, Boston, MA, USA) rabbit antibodies. EGFP-Pdcd4 and endogenous Pdcd4 protein bands are marked by an asterisk and by an arrow, respectively. (C) Evaluation of relative Pdcd4 and eIF4A1 protein abundances in non-cancerous HBEpC cells. Lysate of HBEpC cells (EACCC #502-05a) infected with recombinant replication-deficient adenovirus for EGFP-Pdcd4 protein production (constructed using Ad-Easy system (Stratagene, La Jolla, CA, USA) with derived from pEGFP-Pdcd4 plasmid expression module consisting from CMV promoter-driven EGFP-Pdcd4 cDNA with SV40 virus transcription termination and poyadenylation signal) was incubated with anti-EGFP monoclonal antibody-coated beads. After incubation, bead washing and protein elution from beads as described in (A), proportional amounts of initial lysates (lanes 1), eluted proteins (lanes 2) and flow-through (lanes 3) were analyzed by Western blotting with anti-Pdcd421 or anti-eIF4A1 (Cell Signaling, Boston, MA, USA; Cat. #2490) rabbit antibodies. EGFP-Pdcd4 protein band is marked by an asterisk.](/cms/asset/7b602048-49fe-4ca2-bac3-1b15d8cfa3db/kccy_a_1371881_f0001_b.gif)
Figure 2. eIF4A is not substantially up-regulated in lung cancer cell lines compared to primary human bronchial epithelial cells HBEpC. eIF4A (eIF4A) and eIF4A1 (eIF4A1) proteins were detected by Western blotting with the respective antibodies (Cell Signaling; Cat. #2013 and Cat. #2490, respectively) in total cell lysates of indicated lung cancer cell lines or HBEpC cells. Upper panel shows level of Pdcd4 protein in the lysates of the indicated cell lines detected with anti-Pdcd421 antibodies (Pdcd4). The membrane was also probed with DM1α monoclonal anti-α-tubulin antibodies (Sigma, St. Louis, MO, USA; Cat. #T6199) (α-tubulin) to monitor total protein loading.
Our results rule out a model of Pdcd4 action consisting from mechanistical exhaustion of translation-competent eIF4A isoforms (eIF4A1 and eIFA2). Pdcd4-free eIF4A1 (and, similarly, eIF4A2) would efficiently compete with substantially less abundant eIF4A1:Pdcd4 complex for target mRNA binding thus maintaining eIF4A-dependent translation without noticeable impact of Pdcd4 on it, which is although not the case. As eIF4A proteins reside in the cytoplasm, cytoplasmic Pdcd4 would be almost completely bound by over-representing eIF4A1. Therefore, to be translationally silenced by Pdcd4, mRNA should be transported from the nucleus into the cytoplasm when already in complex with Pdcd4, and only such a pre-complexing, rather than target transcript binding by Pdcd4 in cytoplasm, seems to be permissive for Pdcd4-dependent inhibition of translation.
Indeed, previous experimental results indirectly support this concept. First, Pdcd4 was reported to bind target mRNA in a sequence-specific (likely defined by secondary structure) mode.Citation16,Citation17 Second, Pdcd4 shuttles between cytoplasm and nucleus,Citation22 and nuclear depletion of Pdcd4, which would preclude from inhibition of translation from target mRNA according to the suggested model, was reported to correlate with cancer progression.Citation23-26
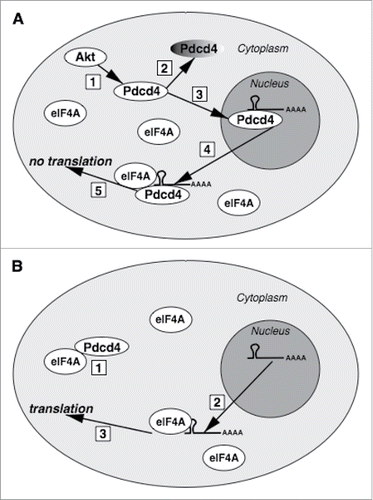
In summary, the above considerations allow us suggesting a novel mechanism of Pdcd4 action as a translation inhibitor and tumor suppressor. The model assumes initial interaction between Pdcd4 and its target mRNAs in the nucleus in a sequence (or structure)-specific manner,Citation18 with subsequent translocation of the Pdcd4:mRNA complex from the nucleus to the cytoplasm, with the translation from such mRNAs being suppressed owing to bound Pdcd4 which blocks helicase activity of eIF4A upon its recruitment on the mRNA. Interestingly, activation of pro-oncogenic Akt signaling has two effects on Pdcd4. First one is Ser67 phosphorylation which primes Pdcd4 for ubiquitin-dependent proteasomal degradation.Citation7 But the second Akt-catalyzed modification, phosphorylation of Ser457, results in protein translocation from the cytoplasm to the nucleus,Citation27 which would protect Pdcd4 from degradation as proteasomes function almost exclusively in the cytosol.Citation28 Therefore, upon pro-oncogenic Akt activation which should be counteracted by Pdcd4 tumor suppressor, only nuclear Pdcd4 would escape degradation, and, according to the proposed model, would retain a capacity to suppress translation from its specific mRNA targets through their primary binding in the nucleus ().
Figure 3. The proposed model of Pdcd4 action. (A) Upon Akt signaling activation (1), Pdcd4 will be targeted for proteasomal degradation (2) and translocated into the nucleus (3). In the nucleus, Pdcd4 can bind its mRNA targets. If translocated from the nucleus to the cytoplasm (4) which might be triggered by unknown stimuli, Pdcd4:mRNA complex would bind eIF4A but, despite abundant eIF4A in the cytoplasm, initial complex formation between Pdcd4 and its target mRNA in the nucleus would preclude from translation (5). (B) Due to excess of eIF4A over Pdcd4, cytoplasmic Pdcd4 would be sequestered by eIF4A (1), with the remaining abundant pool of eIF4A protein capable to support translation from Pdcd4 target mRNAs (3) exported from the nucleus (2).
The suggested concept steaming out from our experimental results and supported by the previous findings, warrants and, most importantly, provides a ground to expedite research aiming to identify specific mRNA targets for Pdcd4 which currently remain mostly unknown, thus contributing to understanding of fundamental mechanisms of Pdcd4 activity as a tumor suppressor and its mechanism of action.
Abbreviations
| EGFP | = | enhanced green fluorescent protein |
| eIF | = | eukaryotic translation initiation factor |
| HBEpC | = | human bronchial epithelial cells |
| MALDI-TOF MS | = | matrix-assisted laser desorption/ionization-time of flight mass-spectrometry |
| Pdcd4 | = | programmed cell death 4. |
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Acknowledgments
This work was carried out using infrastructure of the Center for collective use “Biology of living cell and drug biomedical nanotransporters” of the Institute of Gene Biology, Russian Academy of Science. Authors thank Dr. Denis Logunov and Dr. Maxim Shmarov (N.F. Gamaleya Scientific Research Institute of Epidemiology and Microbiology, Moscow, Russia) for production of recombinant adenovirus for EGFP-Pdcd4 protein expression, and Dr. Rustam Ziganshin (M.M. Shemyakin and Yu.A. Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia) for mass-spectrometric analysis.
Funding
The work was supported by the Russian Foundation for Basic Research under Grants 16-04-00376 and 11-04-00506.
References
- Lankat-Buttgereit B, Göke R. The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol Cell. 2009;101:309-17. doi:10.1042/BC20080191. PMID:19356152
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23:26-37. doi:10.1128/MCB.23.1.26-37.2003. PMID:12482958
- Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol 2004;24:3894-906. doi:10.1128/MCB.24.9.3894-3906.2004. PMID:15082783.
- LaRonde-LeBlanc N, Santhanam AN, Baker AR, Wlodawer A, Colburn NH. Structural basis for inhibition of translation by the tumor suppressor Pdcd4. Mol Cell Biol. 2007;27:147-56. doi:10.1128/MCB.00867-06. PMID:17060447
- Loh PG, Yang HS, Walsh MA, Wang Q, Wang X, Cheng Z, Liu D, Song H. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 2009;28:274-85. doi:10.1038/emboj.2008.278. PMID:19153607
- Suzuki C, Garces RG, Edmonds KA, Hiller S, Hyberts SG, Marintchev A, Wagner G. PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc Natl Acad Sci USA. 2008;105:3274-9. doi:10.1073/pnas.0712235105. PMID:18296639
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467-71. doi:10.1126/science.1130276. PMID:17053147
- Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res. 2008;68:1254-60. doi:10.1158/0008-5472.CAN-07-1719. PMID:18296647
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373-9. doi:10.1038/onc.2008.72. PMID:18372920
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350-9. doi:10.1038/cr.2008.24. PMID:18270520
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-36. doi:10.1038/sj.onc.1210856. PMID:17968323
- Vikhreva PN, Shepelev MV, Korobko IV. mTOR-dependent transcriptional repression of Pdcd4 tumor suppressor in lung cancer cells. Biochim Biophys Acta. 2014;1839:43-9. doi:10.1016/j.bbagrm.2013.12.001. PMID:24334141
- Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, Miller J, Gräf S, Provenzano E, Blows F, et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015;6:e1603. doi:10.1038/cddis.2014.542. PMID:25611378
- Chu J, Pelletier J. Targeting the eIF4A RNA helicase as an anti-neoplastic approach. Biochim Biophys Acta. 2015;1849:781-91. doi:10.1016/j.bbagrm.2014.09.006. PMID:25234619
- Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, Fanidi A. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15:476. doi:10.1186/s13059-014-0476-1. PMID:25273840
- Biyanee A, Ohnheiser J, Singh P, Klempnauer KH. A novel mechanism for the control of translation of specific mRNAs by tumor suppressor protein Pdcd4: inhibition of translation elongation. Oncogene. 2015;34:1384-92. doi:10.1038/onc.2014.83. PMID:24681950
- Biyanee A, Singh P, Klempnauer KH. Translation, Pdcd4 and eIF4A. Oncoscience. 2015;2:731-2. PMID:26501070
- Singh P, Wedeken L, Waters LC, Carr MD, Klempnauer KH. Pdcd4 directly binds the coding region of c-myb mRNA and suppresses its translation. Oncogene. 2011;30:4864-73. doi:10.1038/onc.2011.202. PMID:21643008
- Lankat-Buttgereit B, Göke R. Programmed cell death protein 4 (pdcd4): a novel target for antineoplastic therapy? Biol Cell. 2003;95:515-9. doi:10.1016/j.biolcel.2003.09.003. PMID:14630388
- Raza F, Waldron JA, Quesne JL. Translational dysregulation in cancer: eIF4A isoforms and sequence determinants of eIF4A dependence. Biochem Soc Trans. 2015;43:1227-33. doi:10.1042/BST20150163. PMID:26614665
- Kalinichenko SV, Kopantzev EP, Korobko EV, Palgova IV, Zavalishina LE, Bateva MV, Petrov AN, Frank GA, Sverdlov ED, Korobko IV. Pdcd4 protein and mRNA level alterations do not correlate in human lung tumors. Lung Cancer. 2008;62:173-80. doi:10.1016/j.lungcan.2008.03.022. PMID:18457901
- Böhm M, Sawicka K, Siebrasse JP, Brehmer-Fastnacht A, Peters R, Klempnauer KH. The transformation suppressor protein Pdcd4 shuttles between nucleus and cytoplasm and binds RNA. Oncogene. 2003;22:4905-10. doi:10.1038/sj.onc.1206710. PMID:12894233
- Lim SC, Hong R. Programmed cell death 4 (Pdcd4) expression in colorectal adenocarcinoma: Association with clinical stage. Oncol Lett. 2011;2:1053-7. PMID:23049623
- Kakimoto T, Shiraishi R, Iwakiri R, Fujimoto K, Takahashi H, Hamajima H, Mizuta T, Ideguchi H, Toda S, Kitajima Y, et al. Expression patterns of the tumor suppressor PDCD4 and correlation with β-catenin expression in gastric cancers. Oncol Rep. 2011;26:1385-92. PMID:21894439
- Fassan M, Pizzi M, Giacomelli L, Mescoli C, Ludwig K, Pucciarelli S, Rugge M. PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch. 2011;458:413-9. doi:10.1007/s00428-011-1046-5. PMID:21279518
- Mudduluru G, Medved F, Grobholz R, Jost C, Gruber A, Leupold JH, Post S, Jansen A, Colburn NH, Allgayer H. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer. 2007;110:1697-707. doi:10.1002/cncr.22983. PMID:17849461
- Palamarchuk A, Efanov A, Maximov V, Aqeilan RI, Croce CM, Pekarsky Y. Akt phosphorylates and regulates Pdcd4 tumor suppressor protein. Cancer Res. 2005;65:11282-6. doi:10.1158/0008-5472.CAN-05-3469. PMID:16357133
- Dang FW, Chen L, Madura K. Catalytically Active Proteasomes Function Predominantly in the Cytosol. J Biol Chem. 2016;291:18765-77. doi:10.1074/jbc.M115.712406. PMID:27417138
