ABSTRACT
Shugoshin is an evolutionarily conserved protein, which is involved in tension sensing on mitotic chromosomes, kinetochore biorientation, and protection of centromeric (CEN) cohesin for faithful chromosome segregation. Interaction of the C-terminus of Sgo1 with phosphorylated histone H2A regulates its association with CEN and pericentromeric (peri-CEN) chromatin, whereas mutations in histone H3 selectively compromise the association of Sgo1 with peri-CEN but not CEN chromatin. Given that histone H3 is absent from CEN and is replaced by a histone H3 variant CENP-ACse4, we investigated if CENP-ACse4 interacts with Sgo1 and promotes its association with the CEN chromatin. In this study, we found that Sgo1 interacts with CENP-ACse4 in vivo and in vitro. The N-terminus coiled-coil domain of Sgo1 without the C-terminus (sgo1-NT) is sufficient for its interaction with CENP-ACse4, association with CEN but not the peri-CEN, and this CEN association is cell cycle dependent with maximum enrichment in mitosis. In agreement with the role of CENP-ACse4 in CEN maintenance of Sgo1, depletion of CENP-ACse4 results in the loss of Sgo1 and sgo1-NT from the CEN chromatin. The N-terminus of Sgo1 is required for genome stability as a mutant lacking the N-terminus (sgo1-CT) exhibits increased chromosome missegregation when compared to a sgo1-NT mutant. In summary, our results define a novel role for the N-terminus of Sgo1 in CENP-ACse4 mediated recruitment of Sgo1 to CEN chromatin for faithful chromosome segregation.
Introduction
Erroneous chromosome segregation causes chromosomal instability (CIN), which contributes to aneuploidy observed in several cancers, developmental disorders, and birth defects.Citation1–3 Checkpoints such as the spindle assembly checkpoint (SAC) serve as a quality control mechanism to block the progression of the cell cycle from metaphase to anaphase and prevent CIN.Citation4,5 The SAC monitors kinetochore-spindle attachment and tension between sister chromatids.Citation5–7 The chromosome passenger complex (CPC) which contains the evolutionarily conserved aurora kinase Ipl1 (aurora B), Sli15, Bir1, and Nbl18 responds to erroneous kinetochore-spindle attachments.Citation8 The CPC is recruited to the kinetochore microtubule interface and phosphorylates its substrates thereby allowing for error correction and cell cycle progression.Citation8
In addition to the CPC, evolutionarily conserved Shugoshin (Sgo1 in yeast) also plays a role in sensing kinetochore tension.Citation9,10 The molecular function of shugoshin during meiosis is well established and is evolutionarily conserved among eukaryotic organisms.Citation9,11 One of the functions of Sgo1 is to protect sister-chromatid cohesion duing meiosis I.Citation12–15 Protection of cohesion is achieved by recruitment of heterotrimeric protein phosphatase PP2A to the centromeric chromatin mediated by the N-terminal coiled-coil domain of Sgo1 during meiosis.Citation11,12,15–17 In addition to cohesin protection, shugoshins are also involved in biorientation of homologous chromosomes and spindle checkpoint silencing during meiosis.Citation9,11 In contrast to a well-defined role of Sgo1 in meiosis, there are fewer studies describing a role for Sgo1 in mitosis. In fission yeast, Sgo1 affects centromeric (CEN) localization of Ipl1 kinase during mitosis,Citation10,18 and Xenopus egg extracts depeleted of Sgo1 show altered aurora B kinase activity.Citation19 In budding yeast, Sgo1 has a limited role in cohesion protection in mitosis,Citation20–22 instead, it is involved in tension sensing on mitotic chromosomes, and regulation of kinetochore biorientation.Citation20,21,23,24
Sgo1 associates with core CEN and peri-CEN (50 kb DNA flanking the CEN) regions in budding yeast as well as in other eukaryotic organisms.Citation9,21,23,25 Phosphorylation of histone H2A (S121) by the Bub1 kinase contributes to the recruitment of Sgo1 at the core CEN and peri-CEN regions.Citation24,26,27 The Bub1-H2A-shugoshin mitotic pathway is evolutionarily conserved from budding yeast to mammalian systems.Citation28–31 In addition, the mitotic kinase, Mps1 has been shown to act as an upstream regulator of Sgo1, and CEN recruitment of Sgo1 requires Mps1 kinase activity in budding yeast and human cells.Citation32,33
In meiosis, deletions of kinetochore genes IML3 and CHL4 selectively compromise the association of Sgo1 with peri-CEN but not the core CEN.Citation21 Studies with mitotic cells have shown that an interaction of histone H3 with Sgo1 is required for the association of Sgo1 with peri-CEN but not the core CEN, and that the H3-Sgo1 interaction is essential for tension sensing, and kinetochore biorientation.Citation23 However, histone H3 is not present in budding yeast CEN nucleosomes,Citation34,35 hence additional recruitment mechanisms must regulate the localization of Sgo1 to the core CEN.
Budding yeast contains “point” CENs that are small in size with a well-defined conserved 125 bp DNA sequence in contrast to regional CENs in other eukaryotes, which are composed of alpha-satellite DNA repeats ranging in size from 0.1 to 5 Mb.Citation34,36,37 Despite the variations in CEN DNA, all eukaryotic centromeres contain a specific histone H3 variant (CENP-A in humans, Cse4 in budding yeast) that replaces histone H3 in the CEN nucleosomes.Citation38–40 Unlike regional CENs where CENP-A nucleosomes are interspersed with histone H3 nucleosomes,Citation38,39 point CENs contain only Cse4 nucleosomes, and are devoid of histone H3 nucleosomes making budding yeast an ideal model system to investigate the differences between Sgo1 at the peri-CEN and core CEN. Hence, we investigated the in vivo interaction of Sgo1 with CENP-ACse4, defined the domain of Sgo1 that interacts with CENP-ACse4, and identified the minimum domain of Sgo1 that regulates its association with the core CEN chromatin for faithful chromosome segregation.
In this study, we show that Sgo1 interacts with CENP-ACse4 in vivo and in vitro. The N-terminus coiled-coil domain of Sgo1 (termed as sgo1-NT; amino acid residues 1–150) is sufficient for its interaction with CENP-ACse4. Full length Sgo1 and sgo1-NT associate with CEN chromatin and their CEN association is regulated by the cell cycle with maximum enrichment observed in mitotic cells. While Sgo1 associates with the core CEN and peri-CEN regions, sgo1-NT only associates with the core CEN. CENP-ACse4 is required for the association of Sgo1 and sgo1-NT with core CENs as depletion of CENP-ACse4 results in the loss of Sgo1 and sgo1-NT from the CEN chromatin. Moreover, we found that strains with deletion of the N-terminus of Sgo1 (sgo1-CT) exhibit errors in chromosome segregation. Our results have uncovered a novel interaction of the N-terminus of Sgo1 with CENP-ACse4 that is required for faithful chromosome segregation.
Results
A genome-wide two-hybrid screen with CENP-ACse4 identified Sgo1 as an interacting protein
We performed a genome-wide yeast two-hybrid screen using full length CENP-ACse4 to identify possible interactors of CENP-ACse4 (). CENP-ACse4 was fused to the GAL4 DNA-binding domain (DBD) and this bait strain was screened against the genome-wide array of 6000 prey strains each of these expresses a gene fusion of the open reading frame to the GAL4 activation domain (AD)Citation41 and a mini-array of kinetochore and spindle strains.Citation42 This genome-wide set of two-hybrid strains has successfully been used to identify novel kinetochore protein-protein interactions.Citation42,43 The screen identified ten proteins that interact with CENP-ACse4 (). The Gene Ontology (GO) enrichments for biological process and cellular components for these proteins are available in the supplementary information (Table S1). Included amongst these are two kinetochore proteins, Scm3, Mif2, and histone H4, a constituent of the CENP-ACse4 nucleosomes.Citation44 The identification of pertinent positives that are involved in kinetochore function (p-value = 2E-04; Table S1) validates the two-hybrid screen. Phosphatase (Pph21), and several inter-related kinases including two members of the Snf1 kinase complex (Snf1 and Snf4), its upstream kinase Sak1, as well as Sgo1 were also identified. Even though phosphorylation of CENP-ACse4 has been previously reported,Citation45 a role for Snf1 kinase complex and Sak1 in modification of CENP-ACse4 has not been described so far. We decided to focus on the interaction of Sgo1 with CENP-ACse4 because of the role of Sgo1 in chromosome segregation. We confirmed the two-hybrid interaction using CENP-ACse4 as a DBD-fusion and full length Sgo1 as an AD-fusion. A positive interaction results in the activation of the reporter gene HIS3 and the growth of the transformants on medium lacking histidine with 3mM AT. We observed an interaction of CENP-ACse4 with Sgo1, whereas no growth was observed with the AD vector (pADC), confirming the results of the genome-wide two-hybrid screen ().
Figure 1. Genome-wide two-hybrid screen for CENP-ACse4 identified Sgo1 as an interacting partner. (A) Schematic of yeast two-hybrid assay to identify interactors of CENP-ACse4 used here as a bait. BD = binding domain, AD = activating domain, UAS = upstream activating sequence. GAL4 gene, a transcription factor, produces BD and AD protein products, which are required for transcription of the reporter gene. GAL4-BD+CENP-ACse4 and GAL4-AD+Prey (yeast genes) fusion constructs were used. These fusion proteins alone cannot activate reporter gene transcription; however, expression of both fusion proteins allows interaction between CENP-ACse4 and prey protein leading to transcription of the reporter gene. (B) List of genes showing statistically significant two-hybrid interaction with CENP-ACse4.
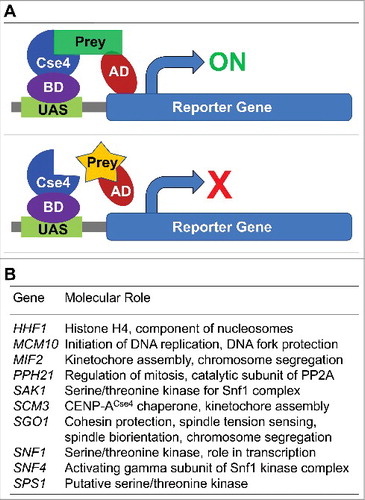
Figure 2. CENP-ACse4 interacts with Sgo1 and sgo1-NT in vivo and in vitro. (A) Sgo1 prey constructs was tested against the CENP-ACse4 bait construct. Sgo1 prey resulted in increased growth compared with vector (pADC) or CENP-ACse4 prey when tested against the CENP-ACse4 FL bait construct. (B) CENP-ACse4 interacts in vivo with Sgo1 and sgo1-NT. Wild type strain (SGO1-MYC, YMB10165), sgo1-NT-MYC (YMB10160) expressing CENP-ACse4 from galactose inducible promoter (GAL1) were grown in YEP with 2% galactose + 2% raffinose at 25°C. Untagged strain (OCF1533-4B), SGO1-MYC (AMY905), and sgo1-NT-MYC (YMB10076) were used as a control. Cell extracts were prepared for immunoprecipitation experiments using anti-HA agarose antibodies (A2095, Sigma-Aldrich). Eluted proteins were analyzed by Western blotting with anti-Myc (Sgo1-Myc or sgo1-NT-Myc; a-14, sc-789, Santa Cruz Biotechnology), anti-HA (CENP-ACse4; H6908, Sigma Aldrich), and anti-Tub2 (loading control) antibodies. IN = input, and IP = immunoprecipitated samples. (C) A minimal prey construct of AD-sgo1-NT132 (amino acids 1–132) resulted in two-hybrid positive interactions with CENP-ACse4-DBD, sgo1-NT (amino acids 1–150) and Rts1-DBD. Scm3-AD was also positive with CENP-ACse4-DBD. Two-hybrid selection plates were SD–HLT and supplemented with 3-aminotriazole (1 or 3 mM). (D) Affinity pull-down of recombinant proteins shows that GST-sgo1-NT132 immobilized on glutathione beads can interact and pull-down His6-CENP-ACse4 octasome from E. coli lysate. IN = input (1% of the input run on a separate gel). *Represents degraded or proteolyzed products. Pull-down experiments were performed three times with similar results.
CENP-ACse4 interacts in vivo with Sgo1 and its N-terminus coiled-coil domain
The identification of Sgo1 as a CENP-ACse4 interacting protein was intriguing and hence we pursued further studies to investigate the biological relevance of this interaction. We performed immunoprecipitation (IP) experiments using agarose beads conjugated with anti-hemagglutinin (HA) antibodies as described in the materials and methods to examine if CENP-ACse4 interacts with Sgo1 in vivo using a wild type strain that expresses Myc-tagged Sgo1 from its endogenous promoter, and HA-tagged CENP-ACse4 from a GAL1 promoter. IP results showed that Sgo1 interacts with CENP-ACse4 in vivo. No signals were observed in control experiments performed using an untagged and Myc-tagged Sgo1 strains ().
Budding yeast Sgo1 contains two distinct domains, which are evolutionarily conserved among eukaryotes: the N-terminus coiled-coil domain (sgo1-NT) that mediates homodimerization,Citation46 and interaction with proteins such as Rts1, a PP2A-B′ subunit,Citation47–49 and the C-terminus basic region, which is required for the interaction of Sgo1 with nucleosomes containing phosphorylated H2A.Citation26 Since the role of N-terminus of Sgo1 is not well defined, we investigated if the N-terminal domain of Sgo1 interacts with CENP-ACse4 by performing IP experiments using a sgo1-NT strain carrying Myc-tagged N-terminus amino acid residues 1–150 expressed from its endogenous promoter. Western blot analysis showed an in vivo interaction between sgo1-NT and CENP-ACse4 ().
To further validate the in vivo interaction of the N-terminus of Sgo1 with CENP-ACse4, we analyzed additional N-terminal Sgo1 constructs using the two-hybrid system with different activation domain fusion contexts. A minimal construct of Sgo1 (amino acids 1–132) fused to the N-terminus of the activation domain (AD-sgo1-NT132) was sufficient to interact with CENP-ACse4 (). Fusion of the Sgo1-NT132 to the C-terminus of the activation domain (sgo1-NT132-AD) did not result in an interaction indicating that this fusion context may hinder the interaction. Consistent with homodimerization of Sgo1 N-terminus,Citation46 a positive interaction was observed between Sgo1 bait (amino acids 1–150; sgo1-NT-DBD) and both the Sgo1 preys (AD-sgo1-NT132 and sgo1-NT132-AD). A similar result was observed for Rts1, which is known to interact with the Sgo1 N-terminal domain.Citation48,49 A positive interaction was observed between CENP-ACse4 and Scm3 used as a control (). Taken together, these results provide evidence for an in vivo interaction of Sgo1 with CENP-ACse4 and show that sgo1-NT is sufficient for its interaction with CENP-ACse4.
CENP-ACse4 interacts in vitro with the N-terminus coiled-coil domain of Sgo1
It is possible that a two-hybrid interaction between sgo1-NT and CENP-ACse4 may be dependent on other proteins. Hence, we examined the interaction of CENP-ACse4 and sgo1-NT using affinity purified CENP-ACse4 and sgo1-NT constructs that were expressed in Escherichia coli. We expressed a GST-tagged N-terminus construct of Sgo1 encompassing amino acid residues 1–132 (GST-sgo1-NT132; predicted molecular weight of 41.4 KDa) and immobilized the protein onto glutathione agarose beads. GST-sgo1-NT132 was tested against E. coli lysate containing RGSH6 tagged CENP-ACse4 co-expressed with histones H2A, H2B and H4. Western blot analysis showed that GST-sgo1-NT132 pulled down CENP-ACse4, whereas the GST control failed to detect any CENP-ACse4 (). Taken together, these results show that the N-terminus coiled-coil domain of Sgo1 (sgo1-NT) is sufficient for its in vitro interaction with CENP-ACse4 and does not require other yeast proteins to mediate the interaction.
N-terminus of Sgo1 is sufficient for association with the core CEN
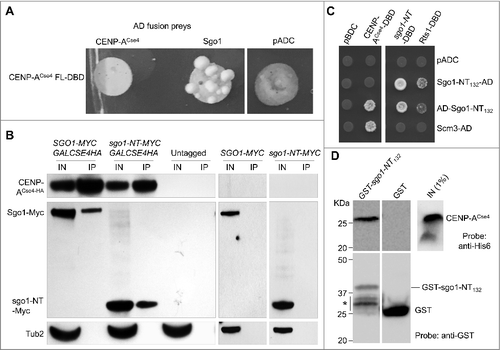
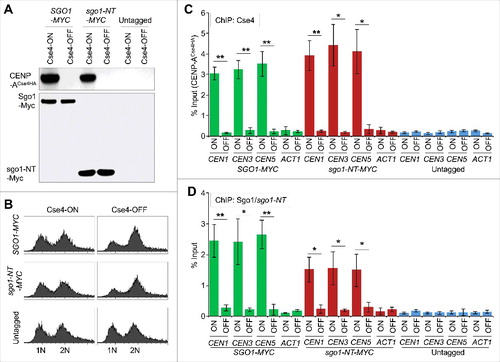
The interaction of Sgo1 and sgo1-NT with CENP-ACse4 in vivo and in vitro prompted us to examine if sgo1-NT can associate with CEN chromatin and if this was cell cycle regulated. Chromatin immunoprecipitation (ChIP) experiments were performed using wild type strains with Myc-tagged Sgo1 or sgo1-NT expressed from its endogenous promoter. Cells were synchronized in G1 (α-factor treatment), S-phase (hydroxyurea treatment), or in G2/M (nocodazole treatment) stages of the cell cycle. Western blot analysis confirmed the protein expression of Sgo1 and sgo1-NT in logarithmically grown cultures and a cell cycle-regulated protein expression of Sgo1 with detectable levels in S and G2/M phases and no detectable level in G1 cells () as reported previously.Citation47 The protein expression level of sgo1-NT was constant throughout the cell cycle, which is consistent with published data indicating that Sgo1 is degraded through a destruction box (amino acids 494–498) in the C-terminal region of the protein.Citation47 The cell cycle synchronization was confirmed by FACS analysis (). Sgo1 enrichment was observed at core CEN (CEN1, CEN3, and CEN5) (, , and ) in logarthimically grown cells consistent with previous studies.Citation21,25 Remarkably, an association of sgo1-NT was also observed at core CEN in logarithmically grown cells (, , and ). No significant enrichment of either Sgo1 or sgo1-NT was detected at the negative control region ACT1 or in an untagged control strain (, , , and ). The enrichment of Sgo1 and sgo1-NT at CEN was significantly higher in mitotic (G2/M) cells, whereas no significant enrichment was observed in G1 and S-phase cells (, and ). Although statistically not significant (p-values = >0.05), the levels of sgo1-NT enrichment at core CEN in G2/M were slightly lower (∼ 29.9 – 36.8%) than those observed for the full length Sgo1 (, , and ). These results show that the N-terminus of Sgo1 associates with the core CEN region in a cell cycle dependent manner independent of its constitutive expression throughout the cell cycle.
Figure 3. Sgo1 and sgo1-NT associate with core CEN in a cell cycle dependent manner. Wild type (SGO1-MYC, AMY905), sgo1-NT-MYC (YMB10076), and untagged control (OCF1533-4B) were grown in YPD to logarithmic phase (LOG) at 25°C, and synchronized in G1 with alpha factor (3 µM), S-phase with HU (0.2 M), and in G2/M with nocodazole (20 µg/mL) at 25°C for 2 hours. ChIP was performed using anti-Myc agarose beads (A7470, Sigma-Aldrich). Enrichment at CEN and at a negative control region ACT1 was determined by qPCR and is shown as % input. Average from three biological replicates ± standard error is shown. **p value <0.01, Student's t test. (A) Western blotting showing expression of Sgo1-Myc and sgo1-NT-Myc in LOG or various stages of the cell cycle. Antibodies used were: anti-Myc (Sgo1 or sgo1-NT; a-14, sc-789, Santa Cruz Biotechnology), and anti-Tub2 (loading control) antibodies. (B) FACS profiles show DNA content representing various stages of the cell cycle. (C) ChIP-qPCR showing enrichment levels of Sgo1-Myc and sgo1-NT-Myc at CEN1 in LOG phase or at various stages of the cell cycle. (D) ChIP-qPCR showing enrichment levels of Sgo1-Myc and sgo1-NT-Myc at CEN3 in LOG phase or at various stages of the cell cycle. (E) ChIP-qPCR showing enrichment levels of Sgo1-Myc and sgo1-NT-Myc at CEN5 in LOG phase or at various stages of the cell cycle. (F) ChIP-qPCR showing enrichment levels of Sgo1-Myc and sgo1-NT-Myc at a negative control region ACT1 in LOG phase or at various stages of the cell cycle.
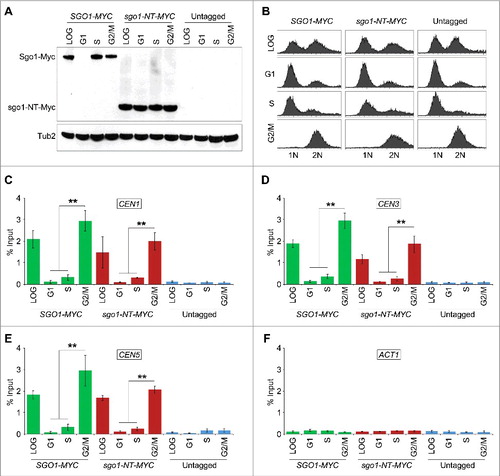
Sgo1 associates with peri-CEN regions, which include about 50 Kb DNA flanking the CEN.Citation9,21 In addition, peri-CEN regions in budding yeast are significantly enriched for cohesin components, Smc3 and Mcd1Citation50,51 consistent with the role of Sgo1 in protection of cohesins.Citation21,52 Hence, we examined whether sgo1-NT associates with peri-CEN regions in G2/M cells (). As expected, Sgo1 associated at peri-CEN regions (). In contrast to Sgo1, sgo1-NT was enriched significantly only at the core CEN but not at peri-CEN regions (). No significant enrichment was detected in an untagged strain used as a negative control ().
Figure 4. sgo1-NT associates with core CEN but not with peri-CEN chromatin. (A) Schematic of CEN and peri-CEN regions on chromosome III. Centromere (CEN3, violet oblong), cohesin associated regions (CARs; green triangle with vertical lines) are shown. CEN is located at 114 kb; peri-CEN regions 115 (1 kb from CEN), 112 (2 kb from CEN), 134 (20 kb from CEN), and chromosome arm CAR 261 (147 kb from CEN) were examined. (B) Association of Sgo1-Myc and sgo1-NT-Myc at CEN and peri-CEN regions. ChIP samples from cells synchronized in G2/M as described in were used. Enrichment at CEN3 and peri-CEN regions was determined by qPCR and is shown as % input. Average from three biological replicates ± standard error is shown.
CEN association of Sgo1 and sgo1-NT is dependent on CENP-ACse4
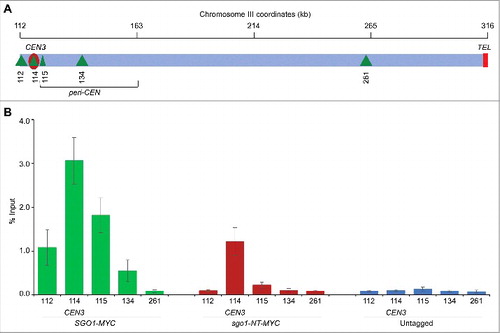
The enrichment of Sgo1 and sgo1-NT at CEN occurs in mitotic cells (), whereas budding yeast CENP-ACse4 is recruited to the CEN in early S-phase and remains associated with CEN chromatin throughout the cell cycle.Citation53,54 Given our results for the association of Sgo1 and sgo1-NT with CENP-ACse4 in vivo (), we posited that CENP-ACse4 may contribute to the maintenance of Sgo1 and sgo1-NT at the CEN. Hence, we constructed strains carrying endogenously expressed Myc-tagged Sgo1 or sgo1-NT in which CENP-ACse4 expression was under the control of galactose inducible promoter (GAL1). CENP-ACse4 is expressed only when these strains are grown in media containing galactose (CENP-ACse4-ON), whereas expression is shut-off when switched to media containing glucose (CENP-ACse4-OFF). ChIP experiments were performed using these strains grown at 25°C in galactose (CENP-ACse4-ON), and after shifting to glucose medium for three hours (CENP-ACse4-OFF). The galactose-induced expression of CENP-ACse4 and glucose mediated shut-off was confirmed by Western blotting in logarithmically growing cultures () and DNA content was determined by FACS analysis (). As expected the enrichment of CENP-ACse4 at core CEN is significantly higher in CENP-ACse4-ON cells, whereas core CEN-associated CENP-ACse4 levels are reduced significantly in CENP-ACse4-OFF conditions (). Sgo1 and sgo1-NT both were significantly enriched at the core CEN under CENP-ACse4-ON conditions. However, the levels of Sgo1 and sgo1-NT were reduced significantly at core CEN in CENP-ACse4-OFF cells (∼10-fold reduction of Sgo1; ∼5-fold reduction of sgo1-NT) (). No significant enrichment of CENP-ACse4, Sgo1, and sgo1-NT was detected at ACT1, or at core CEN and ACT1 regions in an untagged control strain ( and ). These results show that CENP-ACse4 is required for the maintenance of Sgo1 and sgo1-NT at the core CEN chromatin.
Figure 5. Association of Sgo1 and sgo1-NT with core CEN requires CENP-ACse4. Wild type strain (SGO1-MYC, YMB10165), and sgo1-NT-MYC (YMB10160) expressing HA-tagged CENP-ACse4 from galactose inducible promoter (GAL1) were grown in YEP with 2% galactose + 2% raffinose at 25°C (Cse4-ON). Untagged strain (OCF1544-4B) was used as a control. Cells were collected, washed in dH2O and grown at 25°C in YEP with 2% glucose for 3 hours to Shut-off the expression of CENP-ACse4 (Cse4-OFF). Samples were collected for DNA content, protein extraction, and ChIP analyses. (A) CENP-ACse4 protein levels are not detectable upon its depletion. Western blot analysis was carried out on whole cell protein extracts prepared from cultures grown under Cse4-ON and Cse4-OFF conditions as described above. Blots were probed with anti-HA (CENP-ACse4; clone 12CA5, Roche Molecular Systems), and anti-Myc (Sgo1-Myc or sgo1-NT-Myc; a-14, sc-789, Santa Cruz Biotechnology) antibodies. (B) Depletion of CENP-ACse4 causes synchronization of cells in G2/M phase of the cell cycle as revealed by FACS analysis. (C) CEN levels of CENP-ACse4 are reduced upon its depletion. ChIP for CENP-ACse4 was carried out using anti-HA agarose beads (A2095, Sigma-Aldrich). Enrichment at CEN1, CEN3, CEN5, and ACT1 (negative control) was determined by qPCR and is shown as % input. Average from three biological replicates ± standard error is shown. **p value <0.01, *p value <0.05, Student's t test. (D) Sgo1 and sgo1-NT fail to remain associated with CEN chromatin upon depletion of CENP-ACse4. ChIP for Sgo1-Myc and sgo1-NT-Myc was performed using anti-Myc agarose beads (A7470, Sigma-Aldrich). Enrichment at CEN1, CEN3, CEN5, and ACT1 (negative control) was determined by qPCR and is shown as % input. Average from three biological replicates ± standard error is shown. **p value <0.01, *p value <0.05, Student's t test.
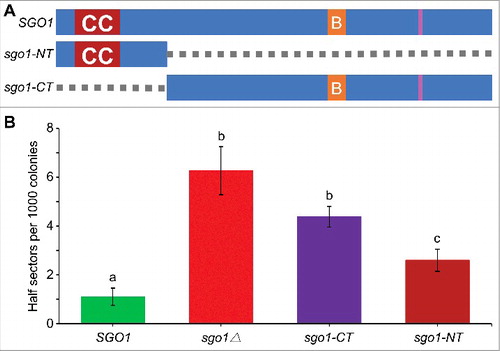
Deletion of N-terminus of Sgo1 (sgo1-CT) causes errors in chromosome segregation
The association of sgo1-NT with core CENs prompted us to examine if the N-terminus of Sgo1 regulates faithful chromosome segregation independent of its C-terminus basic region. A colony color assay was performed to measure the loss of a non-essential reporter chromosome fragment (CF)Citation55 in wild-type, sgo1∆ and sgo1 mutant strains. sgo1-NT expresses Myc-tagged N-terminus amino acid residues 1–150; and sgo1-CT expresses Myc-tagged C-terminus amino acid residues 151–590 (). The frequency of CF loss was measured by counting colonies that were at least half red indicating the loss of CF in the first cell division. The deletion of SGO1 gene results in significantly higher chromosome segregation errors compared to the wild type strain (∼ 6-fold, p-value = 0.008) () as previously reported.Citation27 The frequency of CF loss in sgo1-CT strains is not significantly different than that of sgo1Δ (p-value = 0.153), whereas the frequency of CF loss in sgo1-NT strain is significantly lower than sgo1Δ (p-value = 0.027), and sgo1-CT (p-value = 0.036) strains. The frequency of CF loss in sgo1-NT strain is significantly higher than the wild type strain (∼ 2-fold, p-value = 0.025). Taken together, these results support a role for the N-terminus of Sgo1 in faithful chromosome segregation.
Figure 6. The N-terminus of Sgo1 (sgo1-NT) is required for faithful chromosome segregation. (A) Schematic of full-length SGO1 and its mutant alleles. sgo1-NT includes amino acids residues 1–150 (deletion of amino acids 151–591), whereas, sgo1-CT includes amino acids residues 151–591 (deletion of amino acids 2–150). Symbols: CC denotes the N-terminus coiled coil domain (amino acids 43–87); B denotes the C-terminus basic SGO1 motif (amino acids 366–390), and pink vertical line represents destruction box (amino acids 494–498). (B) sgo1-NT is required for faithful chromosome segregation independently of C-terminus basic domain. Frequency of CF loss in wild type (SGO1; YPH1018), sgo1Δ (YMB10224), sgo1-CT (YMB10221), and sgo1-NT (YMB10225) strains was determined as described in Materials and Methods. At least 1000 colonies from three independent transformants were counted. Average from three biological experiments ± standard error. Values sharing the same letter are not significantly different at a 5% level based on the analysis of variance (p > 0.05).
Discussion
Evolutionarily conserved Sgo1 and its homologs play an important role in cohesin protection, spindle biorientation, and chromosome segregation in eukaryotes.Citation9,13,56–58 Multiple mechanisms regulate the interaction of Sgo1 with nucleosomes, microtubules, and chromatin associated proteins such as cohesin components.Citation9 Phosphorylation of histone H2A by Bub1 kinase regulates the association of Sgo1 to CEN and peri-CEN chromatin.Citation26 Histone H3 interacts with Sgo1 for localization of Sgo1 to peri-CEN in mitosis.Citation23 However, mechanisms that regulate the localization of Sgo1 to the core CEN in mitosis have not been defined. In this study, we have shown that Sgo1 interacts with CENP-ACse4 and the N-terminus of Sgo1 is sufficient for its interaction with CENP-ACse4, and association with core CEN chromatin for faithful chromosome segregation. This conclusion is derived from our observations showing: a) an in vivo and in vitro interaction between Sgo1 and CENP-ACse4 mediated by the N-terminus of Sgo1 (sgo1-NT), b) a cell cycle dependent enrichment of Sgo1 and sgo1-NT to core CENs, c) Sgo1 and sgo1-NT fail to remain associated with core CEN chromatin upon depletion of CENP-ACse4, and d) deletion of N-terminus of Sgo1 (sgo1-CT) causes errors of chromosome segregation independent of its C-terminus basic region.
Our yeast two-hybrid screen for CENP-ACse4 identified genes associated with kinetochore structure and function including Scm3 and Mif2, which have been previously shown to interact with CENP-ACse4 and associate with CEN chromatin.Citation42,59–62 Scm3 is a CENP-ACse4-specific CEN assembly factor, which also protects CENP-ACse4 from ubiquitin mediated proteolysis.Citation60,63–66 Histone H4, a part of the CENP-ACse4 nucleosomesCitation36,44 was also enriched. These results are in agreement with previous in vitro study where CENP-ACse4, Scm3, and histone H4 have been shown to bind each other and form a stoichiometric complex.Citation54,64 In addition to kinetochore genes, we identified new and novel interactors of CENP-ACse4, which are linked with DNA replication and fork protection, Snf1 family kinases, mitotic regulation, and shugoshin.
Our results provide the first evidence of a molecular interaction between CENP-ACse4 and Sgo1 in budding yeast. Sgo1 interacts with CENP-ACse4 in vivo and in vitro and the interaction of CENP-ACse4 with Sgo1 is mediated by the N-terminus coiled coil domain (sgo1-NT), a region of Sgo1 that regulates tension sensing and kinetochore biorientation.Citation47,49,56 A previous study on spatial distribution of budding yeast GFP-tagged proteins at the metaphase kinetochore revealed that CENP-ACse4 and Sgo1 reside in close proximity, where the Sgo1-GFP foci are located 61 nm from CENP-ACse4 toward the inner centromere.Citation28 The proximal fluorescent signals of tagged CENP-ACse4 and Sgo1 suggest potential for an interaction. Moreover, Sgo1 localizes close to the CENP-A signals at metaphase chromosomes in human HeLa cells.Citation26 Furthermore, budding yeast strains with mutations and misregulation of CENP-ACse4 exhibit phenotypes similar to the sgo1Δ strain, for example defects in kinetochore geometry, biorientation and errors in chromosome segregation.Citation28,54,67,68
Sgo1 contains two evolutionarily conserved domains: the N-terminus coiled coil domain, and the C-terminus basic region.Citation9 A recent study has shown that mutations either in the N-terminus coiled coil region or the C-terminus phosphorylated H2A interacting motif of SGO1 exhibit defects in sister chromatid separation and premature SAC silencing.Citation49 We determined that deletion of either N-terminus (sgo1-CT) or C-terminus (sgo1-NT) domains causes increased errors in chromosome segregation indicating that both domains are required and independently regulate chromosome segregation. Our results define a novel role for the N-terminus of Sgo1 in chromosome segregation as the presence of the N-terminus domain (sgo1-NT) reduced the frequency of chromosome loss when compared to strains lacking the N-terminus (sgo1-CT). The higher proficiency of chromosome segregation in the sgo1-NT strain is surprising considering that it only contains a short 150 amino acid N-terminal fragment of Sgo1 that forms a coiled coil based on the crystal structure.Citation46 The N-terminus of Sgo1 mediates homodimerization of Sgo1, which is required for kinetochore biorientation.Citation22,47 Our results suggest that the N-terminus homodimerization domain of Sgo1 alone is sufficient to interact with CENP-ACse4 and has a role in regulating chromosome segregation.
Our results showing expression of sgo1-NT in all phases of the cell cycle is consistent with a stabilized sgo1 mutant (sgo1-Δdb-GFP) that is resistant to APC/C mediated degradation.Citation47 The sgo1-Δdb has the same levels of protein throughout the cell cycle yet exhibits appearance and delocalization timing (sgo1-Δdb-GFP foci) similar to the wild type Sgo1-GFPCitation47 consistent with our results showing lack of CEN localization for sgo1-NT in G1 and S-phases of the cell cycle. It is likely that molecular mechanisms other than Sgo1 protein expression or its destruction through APC/C may regulate its association with the CEN chromatin. Future studies will help us identify molecular factors involved in cell cycle regulated CEN association of Sgo1 in budding yeast.
We determined that the interaction of sgo1-NT with CENP-ACse4 is sufficient for the enrichment of Sgo1 to CEN chromatin. The lower levels of sgo1-NT at core CENs compared to that observed for full length Sgo1 indicates that the C-terminus is also involved in recruitment of Sgo1 to the CEN consistent with studies in budding and fission yeastCitation26 and there is evidence for direct binding to phosphorylated H2A at inner centromeres in human cells.Citation69 The lack of enrichment of sgo1-NT from peri-CEN is supported by the absence of CENP-ACse4 from the peri-CEN chromatin.Citation34,54,70,71 CENP-ACse4 plays a critical role in enrichment and maintenance of Sgo1 and sgo1-NT at the core CEN chromatin. This conclusion is derived from results showing that Sgo1 and sgo1-NT fail to remain associated with the core CEN chromatin upon depletion of CENP-ACse4. We propose that CENP-ACse4, which is recruited to the core CENs in early S-phase and remains there throughout the cell cycleCitation53,54 facilitates the recruitment of Sgo1 specifically to the core CEN only during G2/M. Our results suggest that CENP-ACse4 provides spatial specificity for maintenance of Sgo1 at the core CEN but additional mechanisms promote the dynamic recruitment and dissociation of Sgo1 through the cell cycle. The role of histone H3 in maintenance of Sgo1 at peri-CEN has been described previously,Citation23 and the Bub1-kinase mediated phosphorylation of H2A recruits Sgo1 to peri-CEN and CEN.Citation24,26,27,69
In summary, our results show that CENP-ACse4 interacts with Sgo1, and that this interaction is mediated by the N-terminus coiled coil domain of Sgo1, which regulates association with the core CEN chromatin. We also define a functional role of the N-terminus of Sgo1 in chromosome segregation. Misregulation of CENP-ACse454,Citation72–74 as well as human homologs of Sgo1 (SGOL1 and SGOL2) have been observed in several human cancers.Citation75,76 Hence, future studies aimed at defining how the spatial distribution of Sgo1 at CENs affects chromosome segregation may allow us to determine if misregulation of CENP-ACse4 and Sgo1 contribute to aneuploidy in human cancers.
Materials and methods
Strains, and culture conditions
Budding yeast strains and plasmids are shown in . Strains were grown in yeast peptone dextrose (YPD; 1% yeast extract, 2% Bacto-peptone, 2% glucose), yeast extract peptone (YEP) with 2% galactose + 2% raffinose, or in yeast synthetic medium with 2% glucose.
Table 1. List of yeast strains, plasmids, and primers used in this study.
Chromosome transmission fidelity assay
We used a colony color assayCitation55 to determine the frequency of non-essential chromosome fragment (CF) loss in wild type and sgo1 mutant strains. The loss of CF results in red sectored growth in an otherwise a white colored colony. Strains were grown to logarithmic phase in synthetic glucose medium without histidine to maintain the CF. Cells were collected by centrifugation, dilutions were prepared, and plated on complete synthetic glucose medium with limiting adenine at 25ºC. The frequency of CF loss was determined by counting the number of colonies exhibiting sectors which were at least half red, indicative of CF loss in the first cell division. At least 1000 colonies of three individual transformants were examined for each strain.
ChIP and quantitative PCR (qPCR) analysis
ChIP experiments were performed with three biological replicates using a previously described protocol.Citation54,77 Protein-DNA complexes were captured using anti-Myc agarose (A7470, Sigma-Aldrich, http://www.sigmaaldrich.com/catalog/product/sigma/a7470?lang = en®ion = US) antibodies. ChIP-qPCR was performed using Fast SYBR Green Master Mix (https://www.fishersci.com/shop/products/applied-biosystems-sybr-fast-green-master-mix-6/p-4926995) in 7500 Fast Real Time PCR System (Applied Biosystems) using conditions: 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec, 60°C for 30 sec. The enrichment values were calculated using the ddCT method Citation78 and are shown as % input. Primer sequences are listed in .
IP experiments and Western blotting
IP experiments were performed as described previously.Citation51,63 Briefly, strains were grown to logarithmic phase, and cells were collected by centrifugation. Cell pellets were dissolved in lysis buffer (100 mM Tris [pH 7.5], 0.1 mM EDTA, 150 mM NaCl, 0.1% NP-40, 1 mM DTT, 10% glycerol, and protease inhibitorsCitation63), and whole-cell extracts were prepared by beadbeating using FastPrep-24 5G bead beater (MP Biomedicals, http://www.mpbio.com/product.php?pid = 116005500). Immunoprecipitations were captured using anti-HA agarose antibodies (A2095, Sigma-Aldrich, http://www.sigmaaldrich.com/catalog/product/sigma/a2095?lang = en®ion = US). Beads were washed in the lysis buffer for 5 min at room temperature three times, and eluted in SDS buffer (1% SDS, 1 x TE pH 8.0). Total protein extracts were prepared with TCA procedure and quantified using Bio-Rad DC protein assay (Bio-Rad Laboratories, http://www.bio-rad.com/en-ch/product/dc-protein-assay). Protein samples were analyzed using Western blotting by size fractionation on SDS polyacrylamide gels, followed by transfer to the nitrocellulose membranes. Primary antibodies used were anti-HA (H6908, Sigma-Aldrich, https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Datasheet/2/h6908dat.pdf), anti-HA (clone 12CA5, Roche Molecular Systems, http://www.sigmaaldrich.com/catalog/product/roche/roaha?lang = en®ion = US), anti-Myc (a-14, sc-789, Santa Cruz Biotechnology, https://www.scbt.com/scbt/product/c-myc-antibody-a-14), and anti-Tub254. Secondary antibodies used were HRP-conjugated sheep anti-mouse IgG (NA931V, Amersham Biosciences, http://www.gelifesciences.com/webapp/wcs/stores/servlet/productById/en/GELifeSciences-us/25005173), and HRP-conjugated donkey anti-rabbit IgG (NA934V, Amersham Biosciences, http://www.gelifesciences.com/webapp/wcs/stores/servlet/productById/en/GELifeSciences-us/25005179).
Purification of recombinant proteins from Escherichia coli
Recombinant proteins were purified from E. coli. Expression of GST, GST-Sgo1 and GST-Sgo11-132 were performed in BL21DE3 cells using 100 mL of LB media at 30°C with 3 h induction with 1 mM IPTG (I0328, TCI America, http://www.tcichemicals.com/eshop/en/us/commodity/I0328/). Cells were pelleted, resuspended and lysed with BPER (78248, Thermo Scientific, https://www.thermofisher.com/order/catalog/product/78248) in the presence of protease inhibitor cocktail (88666, Pierce Chemicals, https://www.thermofisher.com/order/catalog/product/88666) and nuclease (Universal nuclease, 88700, Pierce Chemicals, https://www.thermofisher.com/order/catalog/product/88700) as per manufacturer instructions. The CENP-ACse4 octamer was expressed in BL21DE3 cells and kept on ice for 30 min before induction to enhance soluble expression of the CENP-ACse4 octamer. CENP-ACse4 octamer was induced with 0.4 mM IPTG for 3 h. Lysis of the CENP-ACse4 octamer expression strain was perfomed with BPER, protease inhibitor cocktail, nuclease and was supplemented with 2 M NaCl.
In vitro protein-protein interaction assay
The GST and GST-tagged Sgo1 lysates were incubated with 120 µL of glutathione-agarose bead slurry (16101, Pierce Chemicals, https://www.thermofisher.com/order/catalog/product/16101) for 1 h at 4°C and washed five times with equilibration buffer (50 mM Tris-HCl (pH 7.5),150 mM NaCl, 0.1% NP40) and 1 x protease inhibitor (88666, Pierce Chemicals, https://www.thermofisher.com/order/catalog/product/88666). The GST only expression strain lysate was diluted 10 x to ensure a similar level of protein was immobilized on all beads. The GST and GST-Sgo1 protein immobilized beads were incubated for 2 h at 4°C with 1 mL of E. coli CENP-ACse4 octamer expression strain lysate prepared from a 100 mL culture. Beads were washed with 1 mL of equilibration buffer five times. Washed beads were resuspended and boiled at 95°C with 40 μL of SDS-PAGE loading dye. Samples were electrophoresed on 4–20% TGX Stain-Free Gels (Bio-Rad, http://www.bio-rad.com/en-ch/sku/4568095-4-20-mini-protean-tgx-stain-free-protein-gels-12-well-20-ul) and immunblotted with anti-His6 antibody (https://www.thermofisher.com/antibody/product/6x-His-Tag-Antibody-clone-HIS-H8-Monoclonal/MA1-21315) or anti-GST antibody (CGAB-GST, GeneCopoeia, http://www.genecopoeia.com/product/anti-gst-antibody/).
Yeast two-hybrid assay
To screen for CENP-ACse4 protein-protein interactions using yeast two-hybrid, a bait vector was constructed using pBDC and transformed into the haploid strain PJ69-4A MATa.Citation79 This bait strain was then screened against a genome-wide array of prey strainsCitation41 (PJ69-4A MATα) in an automated manner using previously described methods.Citation42,80 To confirm positives, new yeast two-hybrid vectors were constructed and strains were manually retested for positive yeast two-hybrid interactions. Various Cse4, Sgo1 and controls were used as fusion proteins with Gal4 DNA-binding domain (DBD) or Gal4 activation domain (AD) and cloned into the pBDC (TRP1 marked) and pADC (LEU2 marked) vectors.Citation81 The pBDC and pADC vectors were linearized using Nru-I (fusion to the N-terminus of DBD or AD) or Pvu-II (fusion to the C-terminus of DBD or AD) and the gene of interest was PCR amplified and cloned into these vectors using homologous recombination (primers are listed in ). Plasmids were rescued from yeast and confirmed by sequencing. Bait (pBDC) plasmids were transformed into the PJ69-4A MATa strain and prey (pADC) plasmids were transformed into the PJ69-4A MATα haploid strain. Bait and prey strains were mated to form diploid strain by growing the haploid strains in YPAD (Yeast Extract-Peptone-Adenine-Dextrose) medium overnight at 30°C and 50 μL of each strain was mixed in a 96 well plate and diploids were spotted on YPAD plates using a 96 pin spotting tool (VP Scientific, http://www.vp-scientific.com/96_solid_pin_replicator.htm) and incubated overnight at 30°C. To select for the DBD and AD plasmids in diploids, yeast patches were spotted on synthetic dextrose (SD) without tryptophan and leucine (SD-TL) using the 96 pin replicating tool and incubated for 2–3 days at 30°C. Cells from the SD-TL plate were transferred and resuspended into 96 well plates containing 50 μL of SD-TL medium. An aliquot of the yeast diploids were spotted on SD-TL and SD without tryptophan, leucine or histidine (SD-TLH) with 1 mM or 3 mM 3-amino-1,2,4-triazoloe (3-AT, 09540, Fluka, St. Louis, MO) plates and incubated at 30°C for 5–7 days. Positive interactors showed growth on SD-TLH with 3 mM 3-AT.
2017CC7655R-s01.xlsx
Download MS Excel (139.3 KB)Acknowledgments
We are highly thankful to Min-Hao Kuo, Adele Marston, Peter Kaiser, Orna Cohen-Fix, Stefan Milson, and Stan Fields for strains and plasmids; Kathy McKinnon of the National Cancer Institute Vaccine Branch FACS Core for assistance with FACS, and the members of the Basrai laboratory for discussions. PKM, and MAB were supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health; TRH acknowledges support for this work made possible by National Institute of Health grant P30 CA023168 to the Purdue University Center for Cancer Research.
Disclosure statement
The authors declare no competing financial interests.
Additional information
Funding
References
- Pfau SJ, Amon A. A System to Study Aneuploidy In Vivo. Cold Spring Harb Symp Quant Biol. 2015;80:93–101. doi:10.1101/sqb.2015.80.027193. PMID:26936868
- Sansregret L, Swanton C. The Role of Aneuploidy in Cancer Evolution. Cold Spring Harb Perspect Med. 2017;7:1–17. doi:10.1101/cshperspect.a028373. PMID:28049655
- Potapova T, Gorbsky GJ. The Consequences of Chromosome Segregation Errors in Mitosis and Meiosis. Biology (Basel). 2017;6:1–33. PMID:28208750
- Verdaasdonk JS, Gardner R, Stephens AD, Yeh E, Bloom K. Tension-dependent nucleosome remodeling at the pericentromere in yeast. Mol Biol Cell. 2012;23:2560–70. doi:10.1091/mbc.E11-07-0651. PMID:22593210
- Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–224. doi:10.1534/genetics.111.128710. PMID:22491889
- Akiyoshi B, Biggins S. Reconstituting the kinetochore-microtubule interface: what, why, and how. Chromosoma. 2012;121:235–50. doi:10.1007/s00412-012-0362-0. PMID:22289864
- Gimenez-Abian JF, Diaz-Martinez LA, Wirth KG, Andrews CA, Gimenez-Martin G, Clarke DJ. Regulated separation of sister centromeres depends on the spindle assembly checkpoint but not on the anaphase promoting complex/cyclosome. Cell Cycle. 2005;4:1561–75
- Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–40. doi:10.4161/cc.4.11.2146. doi:10.1016/j.tcb.2010.10.007. PMID: 21106376. PMID:16205119.
- Marston AL. Shugoshins: tension-sensitive pericentromeric adaptors safeguarding chromosome segregation. Mol Cell Biol. 2015;35:634–48. doi:10.1128/MCB.01176-14. PMID:25452306
- Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 2007;21:420–35. doi:10.1101/gad.1497307. PMID:17322402
- Jonak K, Zagoriy I, Oz T, Graf P, Rojas J, Mengoli V, Zachariae W. APC/C-Cdc20 mediates deprotection of centromeric cohesin at meiosis II in yeast. Cell Cycle. 2017;16:1145–52. doi:10.1080/15384101.2017.1320628. PMID:28514186
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–7. doi:10.1038/nature02312. PMID:14730319
- Watanabe Y, Kitajima TS. Shugoshin protects cohesin complexes at centromeres. Philos Trans R Soc Lond B Biol Sci. 2005;360:515–21, discussion 21. doi:10.1098/rstb.2004.1607. PMID:15897177
- Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi:10.1016/j.cub.2004.01.051. PMID:14972679
- Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–97. doi:10.1038/nrm1526. PMID:15573136
- Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–72. doi:10.1016/j.cub.2004.03.001. PMID:15062096
- Chambon JP, Touati SA, Berneau S, Cladiere D, Hebras C, Groeme R, McDougall A, Wassmann K. The PP2A inhibitor I2PP2A is essential for sister chromatid segregation in oocyte meiosis II. Curr Biol. 2013;23:485–90. doi:10.1016/j.cub.2013.02.004. PMID:23434280
- Kawashima S, Nakabayashi Y, Matsubara K, Sano N, Enomoto T, Tanaka K, Seki M, Horikoshi M. Global analysis of core histones reveals nucleosomal surfaces required for chromosome bi-orientation. EMBO J. 2011;30:3353–67. doi:10.1038/emboj.2011.241. PMID:21772248
- Rivera T, Ghenoiu C, Rodriguez-Corsino M, Mochida S, Funabiki H, Losada A. Xenopus Shugoshin 2 regulates the spindle assembly pathway mediated by the chromosomal passenger complex. EMBO J. 2012;31:1467–79. doi:10.1038/emboj.2012.4. PMID:22274615
- Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–3. doi:10.1126/science.1101366. PMID:15637284
- Kiburz BM, Reynolds DB, Megee PC, Marston AL, Lee BH, Lee TI, Levine SS, Young RA, Amon A. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 2005;19:3017–30. doi:10.1101/gad.1373005. PMID:16357219
- Peplowska K, Wallek AU, Storchova Z. Sgo1 regulates both condensin and Ipl1/Aurora B to promote chromosome biorientation. PLoS Genet. 2014;10:e1004411. doi:10.1371/journal.pgen.1004411. PMID:24945276
- Luo J, Xu X, Hall H, Hyland EM, Boeke JD, Hazbun T, Kuo MH. Histone h3 exerts a key function in mitotic checkpoint control. Mol Cell Biol. 2010;30:537–49. doi:10.1128/MCB.00980-09. PMID:19917722
- Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr Biol. 2007;17:1837–46. doi:10.1016/j.cub.2007.09.056. PMID:17980598
- Verzijlbergen KF, Nerusheva OO, Kelly D, Kerr A, Clift D, de Lima Alves F, Rappsilber J, Marston AL. Shugoshin biases chromosomes for biorientation through condensin recruitment to the pericentromere. eLife. 2014;3:e01374. doi:10.7554/eLife.01374. PMID:24497542
- Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–7. doi:10.1126/science.1180189. PMID:19965387
- Fernius J, Hardwick KG. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 2007;3:e213. doi:10.1371/journal.pgen.0030213. PMID:18081426
- Haase J, Stephens A, Verdaasdonk J, Yeh E, Bloom K. Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Curr Biol. 2012;22:471–81. doi:10.1016/j.cub.2012.02.006. PMID:22365852
- Liu H, Jia L, Yu H. Phospho-H2A and cohesin specify distinct tension-regulated Sgo1 pools at kinetochores and inner centromeres. Curr Biol. 2013;23:1927–33. doi:10.1016/j.cub.2013.07.078. PMID:24055156
- Asghar A, Lajeunesse A, Dulla K, Combes G, Thebault P, Nigg EA, Elowe S. Bub1 autophosphorylation feeds back to regulate kinetochore docking and promote localized substrate phosphorylation. Nat Commun. 2015;6:8364. doi:10.1038/ncomms9364. PMID:26399325
- Williams SJ, Abrieu A, Losada A. Bub1 targeting to centromeres is sufficient for Sgo1 recruitment in the absence of kinetochores. Chromosoma. 2017;126:279–86. doi:10.1007/s00412-016-0592-7. PMID:27116032
- Storchova Z, Becker JS, Talarek N, Kogelsberger S, Pellman D. Bub1, Sgo1, and Mps1 mediate a distinct pathway for chromosome biorientation in budding yeast. Mol Biol Cell. 2011;22:1473–85. doi:10.1091/mbc.E10-08-0673. PMID:21389114
- van der Waal MS, Saurin AT, Vromans MJ, Vleugel M, Wurzenberger C, Gerlich DW, Medema RH, Kops GJ, Lens SM. Mps1 promotes rapid centromere accumulation of Aurora B. EMBO Rep. 2012;13:847–54. doi:10.1038/embor.2012.93. PMID:22732840
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci U S A. 2007;104:14706–11. doi:10.1073/pnas.0706985104. PMID:17804787
- Melters DP, Nye J, Zhao H, Dalal Y. Chromatin Dynamics in Vivo: A Game of Musical Chairs. Genes (Basel). 2015;6:751–76. doi:10.3390/genes6030751. PMID:26262644
- Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011;12:320–32. doi:10.1038/nrm3107. PMID:21508988
- Buscaino A, Allshire R, Pidoux A. Building centromeres: home sweet home or a nomadic existence? Curr Opin Genet Dev. 2010;20:118–26. doi:10.1016/j.gde.2010.01.006. PMID:20206496
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–21. doi:10.1016/S0092-8674(03)00115-6. PMID:12600307
- Stimpson KM, Sullivan BA. Epigenomics of centromere assembly and function. Curr Opin Cell Biol. 2010;22:772–80. doi:10.1016/j.ceb.2010.07.002. PMID:20675111
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–37. doi:10.1038/nrg2466. PMID:19002142
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi:10.1038/35001009. PMID:10688190
- Wong J, Nakajima Y, Westermann S, Shang C, Kang JS, Goodner C, Houshmand P, Fields S, Chan CS, Drubin D, et al. A protein interaction map of the mitotic spindle. Mol Biol Cell. 2007;18:3800–9. doi:10.1091/mbc.E07-06-0536. PMID:17634282
- Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell. 2003;14:3342–55. doi:10.1091/mbc.E02-11-0765. PMID:12925767
- McKinley KL, Cheeseman IM. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol. 2016;17:16–29. doi:10.1038/nrm.2015.5. PMID:26601620
- Boeckmann L, Takahashi Y, Au WC, Mishra PK, Choy JS, Dawson AR, Szeto MY, Waybright TJ, Heger C, McAndrew C, et al. Phosphorylation of centromeric histone H3 variant regulates chromosome segregation in Saccharomyces cerevisiae. Mol Biol Cell. 2013;24:2034–44. doi:10.1091/mbc.E12-12-0893. PMID:23637466
- Xu Z, Cetin B, Anger M, Cho US, Helmhart W, Nasmyth K, Xu W. Structure and function of the PP2A-shugoshin interaction. Mol Cell. 2009;35:426–41. doi:10.1016/j.molcel.2009.06.031. PMID:19716788
- Eshleman HD, Morgan DO. Sgo1 recruits PP2A to chromosomes to ensure sister chromatid bi-orientation during mitosis. J Cell Sci. 2014;127:4974–83. doi:10.1242/jcs.161273. PMID:25236599
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi:10.1038/nature04664. PMID:16541024
- Jin F, Bokros M, Wang Y. Premature Silencing of the Spindle Assembly Checkpoint Is Prevented by the Bub1-H2A-Sgo1-PP2A Axis in Saccharomyces cerevisiae. Genetics. 2017;205:1169–78. doi:10.1534/genetics.116.195727. PMID:28040741
- Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol. 2000;151:1047–56. doi:10.1083/jcb.151.5.1047. PMID:11086006
- Mishra PK, Ciftci-Yilmaz S, Reynolds D, Au WC, Boeckmann L, Dittman LE, Jowhar Z, Pachpor T, Yeh E, Baker RE, et al. Polo kinase Cdc5 associates with centromeres to facilitate the removal of centromeric cohesin during mitosis. Mol Biol Cell. 2016;27:2286–300. doi:10.1091/mbc.E16-01-0004. PMID:27226485
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi:10.1371/journal.pbio.0030086. PMID:15737064
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–7. doi:10.1016/j.cub.2004.09.086. PMID:15530400
- Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7:e1002303. doi:10.1371/journal.pgen.1002303. PMID:21980305
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–49. PMID:2407610
- Nerusheva OO, Galander S, Fernius J, Kelly D, Marston AL. Tension-dependent removal of pericentromeric shugoshin is an indicator of sister chromosome biorientation. Genes Dev. 2014;28:1291–309. doi:10.1101/gad.240291.114. PMID:24939933
- Kiburz BM, Amon A, Marston AL. Shugoshin promotes sister kinetochore biorientation in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:1199–209. doi:10.1091/mbc.E07-06-0584. PMID:18094053
- Watanabe Y. Shugoshin: guardian spirit at the centromere. Curr Opin Cell Biol. 2005;17:590–5. doi:10.1016/j.ceb.2005.10.003. PMID:16229998
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi:10.1091/mbc.6.7.793. PMID:7579695
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–6. doi:10.1073/pnas.0703178104. PMID:17548816
- Westermann S, Cheeseman IM, Anderson S, Yates JR, 3rd, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–22. doi:10.1083/jcb.200305100. PMID:14581449
- Pinsky BA, Tatsutani SY, Collins KA, Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell. 2003;5:735–45. doi:10.1016/S1534-5807(03)00322-8. PMID:14602074
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–65. doi:10.1016/j.molcel.2007.05.013. PMID:17569568
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–64. doi:10.1016/j.cell.2007.04.026. PMID:17574026
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–54. doi:10.1016/j.molcel.2010.10.014. PMID:21070970
- Mishra PK, Guo J, Dittman LE, Haase J, Yeh E, Bloom K, Basrai MA. Pat1 protects centromere-specific histone H3 variant Cse4 from Psh1-mediated ubiquitination. Mol Biol Cell. 2015;26:2067–79. doi:10.1091/mbc.E14-08-1335. PMID:25833709
- Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics. 2008;179:263–75. doi:10.1534/genetics.108.088518. PMID:18458100
- Collins KA, Camahort R, Seidel C, Gerton JL, Biggins S. The overexpression of a Saccharomyces cerevisiae centromeric histone H3 variant mutant protein leads to a defect in kinetochore biorientation. Genetics. 2007;175:513–25. doi:10.1534/genetics.106.064410. PMID:17151247
- Liu H, Qu Q, Warrington R, Rice A, Cheng N, Yu H. Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Mol Cell. 2015;59:426–36. doi:10.1016/j.molcel.2015.06.018. PMID:26190260
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–13. doi:10.1016/S0092-8674(00)81602-5. PMID:9741625
- Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi:10.1016/j.molcel.2009.07.022. PMID:19782029
- Amato A, Schillaci T, Lentini L, Di Leonardo A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer. 2009;8:119. doi:10.1186/1476-4598-8-119. PMID:20003272
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–6. PMID:12839935
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, Kurumizaka H, Imhof A, Almouzni G. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell. 2014;53:631–44. doi:10.1016/j.molcel.2014.01.018. PMID:24530302
- Kim MS, An CH, Yoo NJ, Lee SH. Frameshift mutations of chromosome cohesion-related genes SGOL1 and PDS5B in gastric and colorectal cancers with high microsatellite instability. Hum Pathol. 2013;44:2234–40. doi:10.1016/j.humpath.2013.04.017. PMID:23850494
- Liu L, Zhang N, Liu J, Min J, Ma N, Liu N, Liu Y, Zhang H. Lentivirus-mediated siRNA interference targeting SGO-1 inhibits human NSCLC cell growth. Tumour Biol. 2012;33:515–21. doi:10.1007/s13277-011-0284-0. PMID:22161216
- Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genetics Genomics. 2007;278:455–65. doi:10.1007/s00438-007-0263-8. PMID:17588175
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–36. PMID:8978031
- Gera JF, Hazbun TR, Fields S. Array-based methods for identifying protein-protein and protein-nucleic acid interactions. Methods Enzymol. 2002;350:499–512. doi:10.1016/S0076-6879(02)50981-2. PMID:12073332
- Millson SH, Truman AW, Piper PW. Vectors for N- or C-terminal positioning of the yeast Gal4p DNA binding or activator domains. Biotechniques. 2003;35:60–4. PMID:12866406
- Choy JS, Acuna R, Au WC, Basrai MA. A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function. Genetics. 2011;189:11–21. doi:10.1534/genetics.111.130781. PMID:21652526
- Mishra PK, Ottmann AR, Basrai MA. Structural integrity of centromeric chromatin and faithful chromosome segregation requires Pat1. Genetics. 2013;195:369–79. doi:10.1534/genetics.113.155291. PMID:23893485
- Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–91. doi:10.1101/gad.1498707. PMID:17242156
- Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell. 2009;20:3818–27. doi:10.1091/mbc.E09-04-0330. PMID:19605555
