ABSTRACT
Accumulating evidence has suggested that circular RNAs (circRNAs) play important roles in oncogenesis and tumor progression. However, our knowledge of circRNAs in gastric cancer (GC) remains limited. To investigate circRNAs involved in GC oncogenesis, we examined differentially-expressed circRNAs and mRNAs in GC tissues and paired noncancerous mucosa tissues using circRNA and mRNA microarrays. Next, we built gene co-expression networks according to the degree of correlation to predict the critical circRNAs in GC. Through bioinformatics analysis, we observed three newly identified circRNAs that are substantially upregulated in GC: hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15. Additionally, hsa_circ_0047905 and hsa_circ_0138960 positively correlated with their parental gene mRNA. Knockdown of hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 in GC cells, resulted in downregulation of parental gene expression. Functional assays suggested that inhibition of these three circular RNAs suppresses GC cell proliferation and invasion in vitro. Those findings suggest that hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 might act as tumor promoters in the pathogenesis of gastric cancer.
Introduction
Gastric cancer (GC) is a common malignant tumor with the fourth-highest incidence, and is the third leading cause of death worldwide.Citation1 According to high-quality data from the National Central Cancer Registry of China, an estimated 697,100 new gastric cancer cases and 498,000 gastric cancer deaths occurred in China in 2015.Citation2 Although the treatment of GC has improved over time, mortality remains high.Citation3 Cancer is widely regarded as a genetic disease, and GC is no exception. However, the molecular and genetic basis of gastric carcinogenesis has not been clearly elucidated. The prognosis of gastric cancer remains poor, in part due to an incomplete understanding of the molecular mechanisms of GC development and progression. Therefore, it is critical to identify new biomarkers and therapeutic targets to improve GC diagnosis and treatment.
Circular RNAs are a novel type of RNAs that form covalently closed continuous loops. The first circRNA was detected in an RNA virus in 1976.Citation4 Three years later, electron microscopic evidence for circular form of RNA was reported in the cytoplasm of eukaryotic cells.Citation5 With the newly-developed technology of next-generation sequencing (NGS), especially RNA-seq technology, more than 30,000 circRNAs have been identified. Owing to their unique structure, they are more stable than linear RNAs. Recent studies have uncovered important functions of circRNAs in multiple biological processes. CircRNAs play many roles, including acting as miRNA spongesCitation6 and regulators of RNA binding protein (RBP) to modulate the expression of protein-encoding genes.Citation7 Some circRNAs even encode peptides.Citation8 It stands to reason, then, that circRNA disorders influence the development of various human diseases, including cancer.Citation9,10 It is important to observe that the unique structure of circRNAs causes them to be insensitive to ribonucleases. Thus, unlike linear RNAs, circRNA molecules can exist in tissues, serum, and urine. Owing to this characteristic, there is tremendous possibility for circRNAs to serve as biomarkers for human cancer.Citation11
The role of circRNAs as molecular markers or potential targets may provide promising application perspectives, such as early tumor diagnosis, therapeutic evaluation, prognosis prediction, and even gene therapy. Several recent studies identified circRNAs that are dysregulated in GC and might be involved in cancer progression.Citation12 Hsa_circ_0001649 was downregulated in GC tissue compared with their paired normal tissues and correlated significantly with pathological differentiation.Citation13 Hsa_circ_0000190 was downregulated in both GC tissues and plasma samples from patients with GC. Its expression levels correlated significantly with tumor diameter, lymphatic metastasis, distal metastasis, TNM stage, and CA19-9 levels. It might be a novel non-invasive biomarker for GC diagnosis.Citation14 Using circRNA microarray chip detection, Zhang et al. analyzed the different expression levels of circRNAs between recurrent GC tissues and non-recurrent GC tissues.Citation15 Another circRNA, circPVT1, which is upregulated frequently in GC tissues because of the amplification of its genomic locus, might promote cell proliferation by serving as a sponge for miR-125 family members.Citation16 Importantly, a growing number of studies have demonstrated that circRNAs are closely associated with disease and may play a significant role in the pathogenesis and diagnosis of disease. However, the characterization and function of circRNAs in human cancer remain largely unknown. Therefore, further study of the mechanisms of interactions between circRNAs and gastric cancer is warranted.
In this study, we investigated various expression profiles of circRNAs and mRNAs between GC tissue and paired noncancerous mucosa tissues with the CapitalBio Technology Human circRNA and mRNA Microarray. We identified a small number of circRNAs that are up- or downregulated in GC tissues compared with normal gastric mucosa tissues. These circRNAs may exhibit an important function in GC and malignancy development and progression. We initially identified three new circRNAs involved in the process of GC development and proposed that these three circRNAs could be used as potential targets in GC therapy.
Results
circRNA and mRNA expression profiles in GC
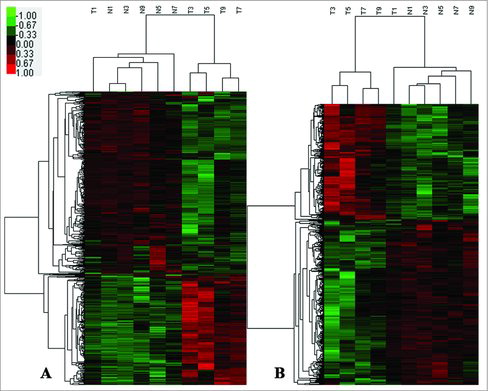
he CapitalBio Technology Human CircRNA Microarray (V2.0, 4 × 180K) and mRNA Human Gene Expression Microarray (V4.0, 4 × 180K) were used to profile circRNA and mRNA expression, respectively, in five paired gastric cancer tissues and adjacent normal tissues (). In total, 170,340 circRNAs and 34,235 coding transcripts (from 21,901 genes) were collected from authoritative databases such as RefSeq, UCSC Known Genes Dataset, Ensembl, Circbase, and Deepbase. To study these microarray data and to re-discover potential biomarkers not yet mined completely, more stringent filtering criteria were used (raw data > 100, fold change > 4, P < 0.05).Citation17 We identified two hundreds and four circRNAs that were differentially-expressed in gastric cancer tissues versus adjacent normal tissues. Seventy-one of these circRNAs were upregulated, and one hundred thirty-three were downregulated, in GC tissues. One hundred sixty-nine mRNAs were differentially-expressed in GC tissues versus paired adjacent normal tissues.
Figure 1. Heat map of the circRNA and mRNA profiles in GC tissues versus paired normal gastric mucosa tissues. Red indicates high relative expression, and green indicates low relative expression. The expression of mRNA (A) and circRNA (B) were hierarchically clustered on the y-axis, and the tissue samples are hierarchically clustered on the x-axis (Fold change ≧ 2, p < 0.05). Expression levels are presented in red and green, indicating upregulated and downregulated RNAs, respectively. Numbers marked with T and N are from five paired gastric cancer tissues and paired adjacent normal tissues, respectively.
Co-expression network to predict critical circRNAs in GC tissues
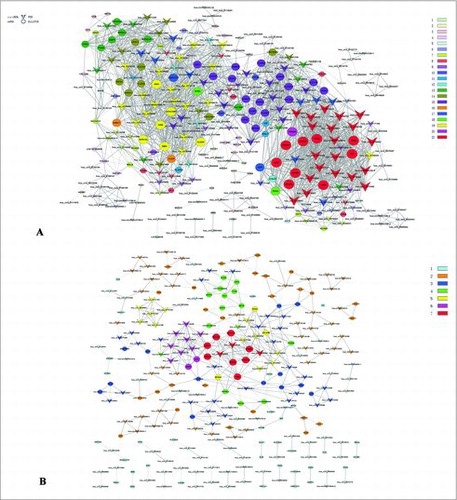
To date, the functions of most circRNAs have not been determined. Therefore, we built a gene co-expression network according to the degree of correlation, in order to predict the critical circRNA in GC tissues.Citation18,19 The structures of the co-expression networks were constructed according to the normalized signal intensities of circRNAs and mRNAs. There are 204 circRNAs and 169 mRNAs in the network analysis. In co-expression networks, each gene corresponds to a node, and 2 genes are connected by a string, indicating a tight correlation between those genes and a potential regulatory relationship (). The co-expression network showed that one circRNA or mRNA correlates with one to dozens of circRNAs or mRNAs. This suggests an interaction between critical circRNAs and mRNAs in GC.
Figure 2. Co-expression network in GC tissues and adjacent normal tissues. The co-expression network consists of 204 circRNAs and 169 mRNAs. (A) GC tissues; (B) Normal tissues. A round node represents a protein-coding gene, and a triangular node represents a circRNA. Solid lines between two nodes indicate positively-correlated interactions between genes, and dashed lines indicate negatively-correlated interactions. Structural cohesion levels are presented in different colors; red indicates a high cohesion level.
Localization of circRNAs in human GC cells and relative expression of circRNAs and parental genes mRNAs in tissues
We posited that circRNAs that interact with more mRNAs targets play more significant roles in the general circRNA-mRNA network than do those that interact with fewer targets. Based on this hypothesis, we designed a step-wise strategy to optimize potential candidate circRNAs. To validate the circRNA microarray and co-expression prediction results, we optimized our parameters to select circRNAs with a microarray raw signal intensity greater than 400 and degree values greater than 20 (Top Degrees = 41). We suggest that the circular RNAs hsa_circ_0047905 (circRNA0047905), hsa_circ_0138960 (circRNA0138960) and has-circRNA7690-15 (circRNA7690-15) () may exhibit important functions in GC and malignancy development and progression. We then designed three sets of divergent primers that amplify only the circular forms, and employed a quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) system to analyze the changes of relative expression. The qRT-PCR results suggest that the circular forms were amplified using the divergent primers, and that expression of hsa_circ_0047905, hsa_circ_0138960, and has-circRNA7690-15 were significantly upregulated in GC tissues compared with noncancerous tissue (, *P < 0.05). This was consistent with microarray data. Fluorescence in situ hybridization (FISH) against circRNA0047905, circRNA0138960 and circRNA7690-15 demonstrated that these three circRNAs preferentially localized in the cytoplasm (, , ). Taken together, these results show that circRNA0047905, circRNA0138960 and circRNA7690-15 are abundant and stable circRNAs expressed in human GC cells.
Figure 3. Localization of circRNAs in AGS human GC cell line. RNA fluorescence in situ hybridization for circRNAs. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Scale bar, 10 μm. (A) circRNA0047905; (B) circRNA0138960; (C) circRNA7690-15.
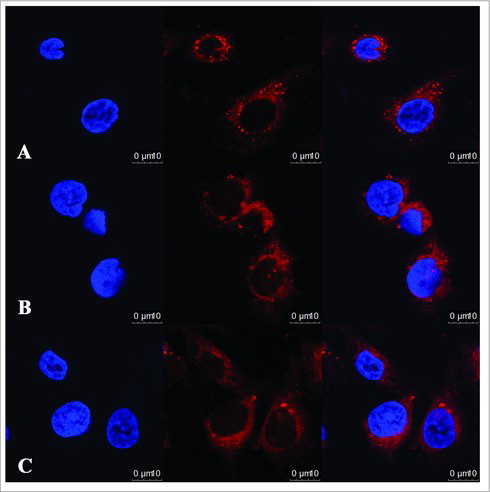
Table 1. Top 3 Critical circRNAs.
To validate the correlation between parental gene linear transcripts (mRNAs) and their circular transcripts (circRNAs), we examined the expression of SERPINB5 and GDA mRNA in 31 pairs of GC tissues and noncancerous tissues. We found that SERPINB5 mRNA was significantly upregulated in GC specimens compared with noncancerous tissues (*P < 0.05). We saw a similar trend for circRNA0047905, but GDA mRNA was not upregulated in GC specimens (P > 0.05, ). We observed a significantly positive correlation between circRNA0047905 and SERPINB5 mRNA in GC tissues (Spearman's correlation, r = 0.7388, *p < 0.0001). We also observed this phenomenon between circRNA0138960 and GDA mRNA (Spearman's correlation, in GC tissues, r = 0.8618, *p < 0.0001). By contrast, there was no positive correlation between circRNA7690-15 and GDA mRNA (Spearman's correlation, in GC tissues, r = 0.03653, p > 0.05, ).
Figure 4. Relative expression of critical circRNAs and parental genes mRNAs in tissues. (A) Relative expression of circRNA0047905, circRNA0138960, circRNA7690-15 in GC tissues compared with paired adjacent noncancerous tissues; (B) Relative expression of SERPINB5 mRNA and GDA mRNA in GC tissues compared with paired adjacent noncancerous tissues (T: GC tissues, N: noncancerous tissue, n = 31). (C) Correlation between expression of circRNAs and parental gene mRNAs in tissue specimens.
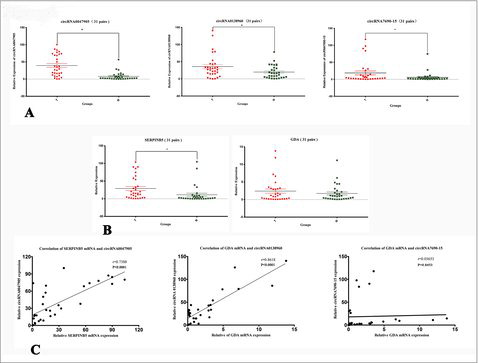
Correlation between circRNA and parental gene mRNA in AGS GC cells
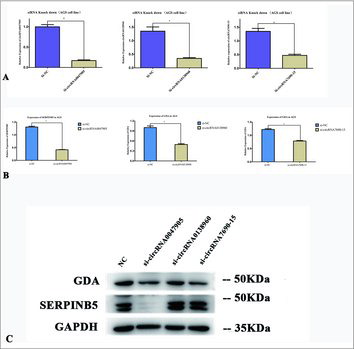
We used small interfering RNA (siRNA) to interfere with the expression of specific genes with complementary nucleotide sequences by degrading circRNAs following transcription. The siRNAs targeted circRNA0047905, circRNA0138960 and circRNA7690-15 at circular RNA back-spliced junction sequences (). Decreased expression of circRNAs in the AGS human gastric cell line was confirmed by qRT-PCR (). Next, we examined parental gene mRNA expression levels for specific siRNA-targeted circRNAs in AGS cells compared with the negative control group. mRNA expression levels of SERPINB5 and GDA () were separately downregulated in the si-circRNA0047905, si-circRNA0138960 and si-circRNA7690-15 cell groups, and protein expression () levels of SERPINB5 were significantly downregulated in the si-circRNA0047905 group. This suggests that circRNA0047905, circRNA0138960 and circRNA7690-15 regulate expression of their parent genes in gastric cancer cells.
Table 2. siRNAs targeted at circRNA backspliced junction sequences.
Figure 5. Relative expression of circRNAs and mRNAs in AGS human GC cell lines transfected with specific siRNA targeted circRNAs. (A) Relative expression of circRNAs in AGS GC cell lines transfected with specific siRNA. *P < 0.05. (B) Relative expression of parental gene mRNAs in AGS GC cell lines transfected with specific siRNAs respectively targeted circRNAs. *P < 0.05. (C) Expression of SERPINB5 and GDA protein in AGS GC cell line transfected with specific siRNAs.
Depletion of circRNA0047905, circRNA0138960 and circRNA7690-15 suppress AGS cell proliferation and invasion in vitro
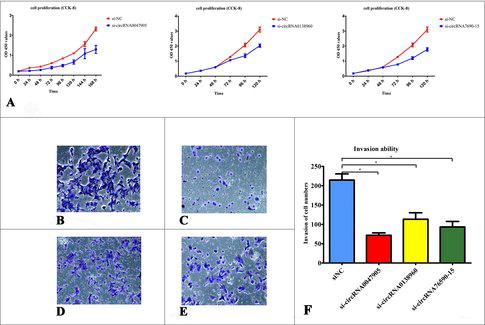
To evaluate the biological effects of circRNA0047905, circRNA0138960 and circRNA7690-15 in GC cells, we compared the proliferation and invasion ability of AGS human gastric cancer cells. CCK-8 assays revealed that cell proliferation was suppressed in cells when transfected with specific siRNA, compared with negative controls (). Furthermore, knockdown of circRNA0047905, circRNA0138960 and circRNA7690-15 resulted in attenuated invasion, as measured by Transwell assays (, , , , ).
Figure 6. Tumor-promoting effects of circRNAs in AGS human GC cell line. (A) Tumor-promoting effects of circRNA0047905, circRNA0138960, circRNA7690-15 in AGS cell lines. The proliferation curve of AGS cells following transfection with specific siRNAs by the CCK8 assay. The effects of circRNA0047905, circRNA0138960, and circRNA7690-15 knockdown on the invasive ability of AGS cells were assessed by the Transwell method. (B) si-Negative control, (C) si-circRNA0047905, (D) si-circRNA0138960, (E) circRNA7690-15), (F) Quantification was performed by light microscope (200 x), counting the stained cells that invaded the lower chamber, *P < 0.05.
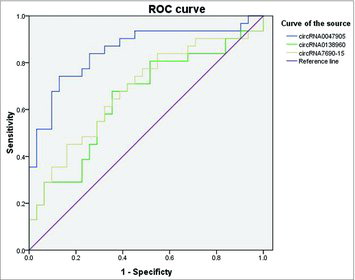
Diagnostic value of circRNA0047905, circRNA0138960 and circRNA7690-15
We further evaluated the diagnostic value of these three circRNAs for GC by plotting a receiver operating characteristic curve (). We found that circRNA0047905 had the highest diagnostic accuracy, with an area under the curve (AUC) of 0.85 (95% CI: 0.751-0.950). circRNA0138960 and circRNA7690-15 had lower diagnostic accuracy. The AUC of circRNA0138960 and circRNA7690-15 were 0.647 (95% CI: 0.508-0.786) and 0.681 (95% CI: 0.546-0.815), respectively.
Discussion
Via high-throughput RNA sequencing and bioinformatics analysis, circRNAs have now been recognized as a large species of RNAs with thousands of identified sequences in animal cells.Citation20-22 CircRNAs often show developmental stage-specific and tissue-specific expression; thus, differentially expressed circRNAs in a wide range of cancers may play important roles in cancer initiation and progression.Citation23 In this study, we performed a microarray assay to explore the aberrant expression profile of circRNAs and mRNAs in GC tissue compared with that of paired noncancerous mucosal tissue. Many circRNAs were significantly differently-expressed, suggesting an important role of circRNAs in GC. Applying a bioinformatics approach, we uncovered three circular RNAs (circRNA0047905, circRNA0138960, and circRNA7690-15) that were significantly upregulated and that may play critical roles in GC. By analyzing RT-PCR experimental data, we confirmed that circRNA0047905, circRNA0138960 and circRNA7690-15 were significantly upregulated in GC tissue. In addition, circRNA0047905 and circRNA0138960 expression levels correlated significantly with parental gene mRNA expression. We suggest that circRNA0047905, circRNA0138960 and circRNA7690-15 might promote AGS cell proliferation and invasion in vitro.
Mammary serine protease inhibitor B5 (SERPINB5) is a 42-kDa protein that is a member of the ovalbumin clade of serine protease inhibitors. SERPINB5 was identified by subtractive hybridization and differential display, and was found to be expressed in normal mammary epithelial cells but not in most mammary carcinoma cell lines, and was considered to be a tumor suppressor.Citation24 However, conflicting reports on its function in gastric cancer occurrence and progression have been reported.Citation25-28 These findings suggest that SERPINB5 may function differently in gastric cancer compared with other malignant tumors. We need further research to reveal the specific biological mechanisms of the biomolecules. This investigation highlighted the importance of circRNA0047905. SERPINB5 is the parental gene of circRNA0047905. Gastric tumor specimens showed an increased SERPINB5 expression level compared with that of corresponding normal tissues. The frequency of SERPINB5 induction was associated with the stage of gastric cancer (GC) and lymph node metastasis.Citation25-28 SERPINB5 may play an important role in the progression and metastasis of gastric adenocarcinoma by acting as an oncogene through its interaction with KHDRBS3 and FBXO32.Citation25-28 In our study, we observed a significant positive correlation between circRNA0047905 and SERPINB5 mRNA in GC tissues. circRNA0047905 had the highest diagnostic accuracy with an area under the curve (AUC) of 0.85 (95% CI: 0.751-0.950). It may prove to be a promising biomarker in GC diagnosis. Further investigations will be required to determine the mechanism, and the clinical significance, of higher SERPINB5 and circRNA0047905 expression in gastric cancer.
Recently, an argument has been raised that only very limited numbers of circRNAs (possibly only a handful) act as microRNA sponges.Citation29,30 The specific RNA-RNA interaction between circRNAs and mRNAs might be one of the central themes underlying the functional mechanism of circRNAs.Citation30,31 According to our results, we speculate that circRNA0047905, circRNA0138960and circRNA7690-15 might modulate expression of their parental genes transcriptionally. The functions and related mechanisms of circRNAs may be rather diverse; therefore, further studies should be performed to rigorously explore the physiological roles of circRNAs.
Conclusion
This study examined co-expression networks between circRNAs and mRNAs, leading us to find critical certain circRNAs in GC. circRNA0047905, circRNA0138960 and circRNA7690-15 were found to be candidate oncogenes in GC. As a promising biomarker in GC tissues, circRNA0047905 had the highest diagnostic accuracy in the current study. We observed a significantly positive correlation between circRNA0047905 and SERPINB5 mRNAs. This phenomenon also existed between circRNA0138960 and GDA mRNA. Finally, our results suggested that circRNA0047905, circRNA0138960 and circRNA7690-15 promote AGS cell proliferation and invasion in vitro.
Materials and methods
Patient samples
Thirty-one GC tissue and paired adjacent noncancerous mucosal tissue samples were obtained from the Department of General Surgery, Peking University Peoples' Hospital. The samples were immediately snap-frozen in liquid nitrogen after resection and were stored at −80°C until RNA extraction. The diagnosis of GC was confirmed pathologically. Five pairs of samples were used for microarray analysis, and thirty-one pairs were used for qRT-PCR validation. All patients provided written informed consent prior to sample collection. The study was approved by the local Research Ethics Committee of Peking University.
Cell lines and cell culture
The human GC cell line AGS was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). It was tested and authenticated for genotypes by DNA fingerprinting. The cell line was passaged for fewer than 6 months after reconstitution. AGS was cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS) (all from Thermo Fisher, USA) at 37°C with 5% CO2.
RNA extraction and purification
Total RNA was extracted from tissue samples using Trizol reagent (Invitrogen, USA) and was purified using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer's protocol. The purity and concentration of RNA were determined from OD260/280 readings using a spectrophotometer (Nano-Drop ND-1000). RNA integrity was determined by 1% formaldehyde denaturing gel electrophoresis.
CircRNA and mRNA microarray expression profiling
Microarray profiling was conducted in the laboratory of Capitalbio Technology Corporation in Beijing, China. Sample labeling, microarray hybridization and washing were performed according to manufacturer's standard protocols. Briefly, each purified RNA sample was transcribed to double-strand cDNA, and was then synthesized into cRNA and labeled with Cy3-dCTP. The labeled cRNAs were hybridized onto the CapitalBio Technology Human CircRNA Microarray v2.0 (V2.0,4 × 180K) and mRNA Human Gene Expression Microarray (V4.0,4 × 180K), including the global profiling of 170,340 circRNAs and 34,235 coding transcripts (from 21,901 genes). Next, the arrays were washed and scanned using the Agilent Scanner G2505C (Agilent Technologies). The circRNA and mRNA array data were analyzed for data summary, normalization and quality control using GeneSpring software V13.0 (Agilent). To select differentially-expressed genes, we used threshold values ≧ 2-fold and ≦0.5-fold change, and a Benjamini-Hochberg corrected p value of 0.05.
Co-expression network
Gene co-expression networks were built according to the normalized signal intensity of specific expression genes. For each pair of genes, we calculated the Pearson correlation and chose the significant correlation pairs to construct the network.Citation32 Within the network analysis, degree centrality is the simplest and most important measure of the centrality of a gene within a network that determines relative importance. Degree centrality is defined as the number of links one node has to the other. To study some properties of the networks, k-cores in graph theory were introduced as a method of simplifying graph topology analysis. A k-core of a network is a subnetwork in which all nodes are connected to at least k other genes in the subnetwork.Citation33 The purpose of Network Structure Analysis is to locate core regulatory factors (genes) in one network.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Reverse transcription was performed using a reverse transcription kit (Invitrogen, USA). The expression of circRNAs and mRNAs was measured by qRT-PCR using the iTaq™ Universal SYBR® Green Supermix (Bio-Rad, USA) in the CFX96 Real-Time PCT Detection System (Bio-Rad, Hercules, CA). The primer sequences are displayed in . Human β-Actin RNA was amplified as an internal control for circRNAs and mRNAs. circRNA and mRNA expression levels were calculated according to the 2−△△Ct method.
Table 3. Primer sequences for qRT-PCR.
RNA fluorescence in situ hybridization (RNA FISH)
Circular RNAs FISH probes were synthesized by Ribo Bio Technology Co Ltd (Guangzhou, China). FISH was performed with the FISH kit according to the manufacturer's protocol. Cells were fixed with 4% paraformaldehyde for 10 min at room temperature, then permeabilized in PBS with 0.5% TritonX-100 on ice for 5 min. This was followed by pretreatment with pre-hybridization buffer at 37°C for 30 min. Subsequently, cells were hybridized with 20 μM Cy3-labeled RNA of circRNAs FISH probe mix in a moist chamber at 37°C overnight. Cells were rinsed three times in 4 × SSC with 0.1% Tween-20 for 5 min at 42°C, followed by washing once for 5 min at 42°C in 2 × SSC, and then were washed once for 5 min at 42°C in 1 × SSC. After hybridization, cells were stained with 6-diamidino-2-phenylindole (DAPI) for 10 min at room temperature. Finally, images were acquired on a Leica SP8 STED confocal microscope (Leica Microsystems, Mannheim, Germany).
Generation of knockdown cell lines
Small interfering RNAs (siRNAs) were synthesized by Ribo Bio Technology Co Ltd (Guangzhou, China) to interfere with the expression of specific genes with complementary nucleotide sequences by degrading circRNAs. siRNAs were transfected with Lipofectamine™ RNAiMAX Transfection Reagent (Invitrogen, USA). The expression of circRNAs and mRNAs in transfected cells were confirmed by qRT-PCR.
Cell proliferation assay
Cells (∼2,500) were seeded onto 96-well plates and were incubated at 37°C after transfection. After incubation for 1 to 6 days, 10 μl of CCK8 solution was added each well, and the plates continued to be incubated for another 1.5 h at 37°C. Absorbance was measured at 450 nm using a microplate reader (Bio-Rad). The CCK8 assay was repeated three times in triplicate.
Matrigel invasion assay
The Matrigel invasion chamber was used to assess cell invasion ability (24-well plates, 8-mm pore size; Corning). Briefly, cells were seeded in the upper chamber with media containing 0.1% bovine serum albumin, while media containing 30% FBS were placed in the lower well. After incubation for 48 h at 37°C, the non-invading cells were removed with cotton swabs. Invasive cells at the bottom of the membrane were fixed with 1% formalin, stained with 0.1% crystal violet and counted under microscopic vision. Invasion detection was repeated three times in duplicate.
Western blot analysis
Total protein from the cells was extracted using cell lysis buffer with proteinase inhibitors (Roche Diagnostics, Basel, Switzerland). Lysates were denatured with sodium dodecyl sulfate (SDS) buffer at 100°C for 10 min, separated in 10% or 15% polyacrylamide gels and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Hertfordshire, UK). The membranes were blocked with 5% non-fat milk powder in Tris-buffered saline containing 0.1% Tween 20 (TBST) and probed with MASPIN (SERPINB5) antibody (ab66513; Abcam plc, Cambridge, UK) and GDA antibody (ab155773; Abcam plc, Cambridge, UK) overnight at 4°C. The following day, the membranes were treated with secondary antibodies. The band signals were visualized through enhanced chemiluminescence (ECL; Pierce, Rockford, IL, USA) after exposure to a ChemiDoc™ XRSC system (Bio-Rad).
Statistical analysis
All results were expressed as means ± SD. Differences between groups were assessed using a paired t-test. P < 0.05 was considered statistically significant. The relationship between the expression of circRNAs and mRNAs was evaluated by Spearman's correlation. P < 0.05 was considered statistically significant. All data, unless otherwise explained specifically, were analyzed using SPSS 20.0 software (SPSS, Chicago, IL, USA).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of our laboratory for helpful discussion and Novel Bioinformatics Ltd., Co for the support of bioinformatics analysis with their NovelBio Cloud Analysis Platform.
Additional information
Funding
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. doi:10.3322/caac.21262. PMID:25651787
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66:115-32. PMID:26808342
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86. doi:10.1002/ijc.29210. PMID:25220842
- Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547-55. doi:10.1016/0092-8674(76)90223-3. PMID:182384
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339-40. doi:10.1038/280339a0. PMID:460409.
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8. doi:10.1038/nature11993. PMID:23446346
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64. doi:10.1038/nsmb.2959. PMID:25664725
- Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859:1245-51. doi:10.1016/j.bbagrm.2016.07.009. PMID:27449861
- Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17:679-92. doi:10.1038/nrg.2016.114. PMID:27739534
- Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53:359-65. doi:10.1136/jmedgenet-2016-103758. PMID:26945092
- Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. doi:10.1186/s12943-017-0598-7.
- Huang G, Li S, Yang N, Zou Y, Zheng D, Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8-18. doi:10.1016/j.canlet.2017.07.002. PMID:28705773
- Li W, Song Y, Zhang H, Zhou Z, Xie X, Zeng Q, Guo K, Wang T, Xia P, Chang D. Decreased expression of Hsa_circ_00001649 in Gastric cancer and its clinical significance. Dis Markers. 2017;2017:1-6. doi:10.1155/2017/7089493.
- Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167-71. doi:10.1016/j.cca.2017.01.025. PMID:28130019
- Zhang Y, Li J, Yu J, Liu H, Shen ZY, Ye GT, Mou TY, Qi XL, Li GX. Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. 2017;8:22936-43.
- Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208-19. doi:10.1016/j.canlet.2016.12.006. PMID:27986464
- Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, et al. LncRNA GClnc1 promotes Gastric Carcinogenesis and may act as a modular Scaffold of WDR5 and KAT2A complexes to specify the Histone modification pattern. Cancer Discov. 2016;6:784-801. doi:10.1158/2159-8290.CD-15-0921. PMID:27147598
- Prieto C, Risueno A, Fontanillo C, De Las RJ. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. Plos One. 2008;3:e3911. doi:10.1371/journal.pone.0003911. PMID:19081792
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101-13. doi:10.1038/nrg1272. PMID:14735121
- Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi:10.1093/nar/gkl151. PMID:16682442
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8. doi:10.1038/nature11928. PMID:23446348
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. Plos Genet. 2013;9:e1003777. doi:10.1371/journal.pgen.1003777. PMID:24039610
- Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53:359-65. doi:10.1136/jmedgenet-2016-103758. PMID:26945092
- Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526-9. doi:10.1126/science.8290962. PMID:8290962
- Kim SM, Cho SJ, Jang WY, Kim DH, Shin HS, Jang MK, Kim HY, Nam ES. Expression of maspin is associated with the intestinal type of gastric adenocarcinoma. Cancer Res Treat. 2005;37:228-32. doi:10.4143/crt.2005.37.4.228. PMID:19956519
- Song SY, Son HJ, Kim MH, Nam ES, Rhee JC, Park C. Prognostic significance of maspin expression in human gastric adenocarcinoma. Hepatogastroenterology. 2007;54:973-6. PMID:17591106
- Yu M, Zheng H, Tsuneyama K, Takahashi H, Nomoto K, Xu H, Takano Y. Paradoxical expression of maspin in gastric carcinomas: correlation with carcinogenesis and progression. Hum Pathol. 2007;38:1248-55. doi:10.1016/j.humpath.2006.11.025. PMID:17490717
- Lei KF, Liu BY, Wang YF, Chen XH, Yu BQ, Guo Y, Zhu ZG. SerpinB5 interacts with KHDRBS3 and FBXO32 in gastric cancer cells. Oncol Rep. 2011;26:1115-20. PMID:21725612
- Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi:10.1186/s13059-014-0409-z. PMID:25070500
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64. doi:10.1038/nsmb.2959. PMID:25664725
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Corrigendum: Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2017;24:194. doi:10.1038/nsmb0217-194a. PMID:28170000
- Prieto C, Risueno A, Fontanillo C, De Las RJ. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. Plos One. 2008;3:e3911. doi:10.1371/journal.pone.0003911. PMID:19081792
- Vazquez A, Dobrin R, Sergi D, Eckmann JP, Oltvai ZN, Barabasi AL. The topological relationship between the large-scale attributes and local interaction patterns of complex networks. Proc Natl Acad Sci U S A. 2004;101:17940-5. doi:10.1073/pnas.0406024101. PMID:15598746