ABSTRACT
Chromosome instability (CIN) contributes to the development of many cancer. In this paper, we summarize our recent finding that a novel pathway by which FBW7 loss promotes Centromere Protein A (CENP-A) phosphorylation on Serine 18 through Cyclin E1/CDK2, therefore promoting CIN and tumorigenesis. Our finding demonstrates the importance of CENP-A post-translational modification on modulating centromere and mitotic functions in cancer.
Chromosome instability (CIN) contributes to tumor heterogeneity, drug resistance and cancer progression, and high levels of CIN are associated with poor patient survival for many cancer types.Citation1,Citation2 Major mechanisms proposed for CIN include oncogene-induced replication stress, telomere dysfunction, and aberrant mitosis.Citation1 FBW7 is a tumor suppressor protein frequently mutated in multiple cancer types, and belongs to the F-box protein family as part of the SCF ubiquitin E3 ligase complex.Citation3 Cyclin E1 is a well-characterized FBW7 substrate that regulates G1/S cell cycle entry. Aberrant accumulation of Cyclin E1 due to overexpression or FBW7 mutation leads to polyploidyCitation4, and is prevalent in many types of cancers. Existing evidence strongly suggests that Cyclin E1 misregulation results in genomic instability primarily through excessive replication origin firing and oncogene-induced replication stress.Citation5 However, we have recently identified a novel pathway where FBW7 modulates phosphorylation of an essential centromere protein CENtromere Protein A (CENP-A) through Cyclin E1/CDK2, presenting a new paradigm for how this tumor suppressor regulates CIN and tumorigenesis through centromere and mitotic functions.Citation6
The centromere is the specialized chromatin locus that recruits the kinetochore, and is crucial for proper mitosis and genome maintenance. The centromere is enriched for CENP-A, an important histone H3 variant that is considered a key epigenetic mark for centromere identity and propagation.Citation7,Citation8 Too little or too much of CENP-A can disrupt genome integrity. CENP-A depletion displaces the downstream components of the Constitutive Centromere-Associated Network (CCAN) and the KMN network (KNL1 complex, MIS12 complex, NDC80 complex) from centromeres and kinetochores, resulting in chromosome missegregation.Citation9-12 Moreover, ectopic localization of CENP-A to non-centromeric loci due to overexpression or targeted recruitment leads to fragmented chromosomes in Drosophila melanogaster and human cell lines.Citation13-16 At least in Drosophila this is through formation of neo-centromeres and ectopic kinetochores.Citation13-16 Therefore, faithful chromosome segregation requires tight regulation of CENP-A protein levels, to ensure proper CENP-A nucleosome assembly only at centromeres.Citation17 In tumors, overexpression of centromere and kinetochore genes is prevalent.Citation18,Citation19 Importantly, centromere gene upregulation strongly correlates with CIN and poor prognosis in numerous human cancer typesCitation18-23, and predicts enhanced cancer cell and patient sensitivity to genotoxic adjuvant therapies.Citation18 However, it is unclear whether centromere misregulation contributes to malignancy, or is merely a consequence of other changes during tumor progression.
Post-translational modifications of centromere proteins also influence centromere functions. These include phosphorylation of Ser16 and Ser18 residues within the CENP-A N terminal tailCitation24, as well as Ser68 in the histone fold domain.Citation25,Citation26 Ser to Ala mutations at these sites lead to defective CENP-A deposition or mitotic defects, although there is some debate about how essential these modifications are in normal cells.Citation26,Citation27 Nevertheless, CENP-A and other centromere and kinetochore protein genes are rarely mutated in large TCGA patient datasetsCitation18, thus the clinical relevance of these modifications and the potential roles of CENP-A regulation in cancer progression remain poorly understood.
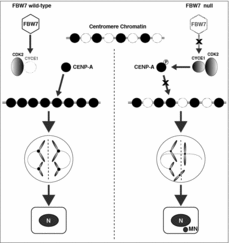
We found that increased Cyclin E1 levels promote CIN and tumor growth through centromere misregulation.Citation6 Specifically, loss of the tumor suppressor FBW7 results in increased Cyclin E1/CDK2 activity, leading to hyper-phosphorylation of CENP-A at the N- terminal Ser18 site, and reduced CENP-A, CENP-B and HEC1 levels at centromeres. Mechanistically, our study demonstrated that Cyclin E1/CDK2 is necessary and sufficient for CENP-A Ser18 phosphorylation both in vitro and in cultured cells. Further, excessive CENP-A Ser18 phosphorylation enhances CIN, including chromosome missegregation and micronucleus formation, and promotes anchorage-independent growth and tumor progression (). Strong evidence for the relevance of their findings to human clinical cancers was demonstrated using a disease relevant FBW7 mutation, human clinical cancer tissues and a xenograft mouse model. Moreover, our results suggest that Ser18 hyper-phosphorylation due to increased Cyclin E1 activity and/or FBW7 loss reduces efficient CENP-A deposition at centromeres. Mechanistically, these results suggest that the HJURP chaperone and assembly factor does not interact with CENP-A S18D (Ser to Asp) mutant as efficiently as with WT proteins. Together with the data from clinical human breast cancer samples, we identify an important new function for aberrant Cyclin E1/CDK2 activation in cancer, distinguishable from its well-established role in the G1/S transition.Citation6 Previous research on Cyclin E1 in oncogenesis primarily focused on its role in replication initiation, which supports oncogene-induced replication stress.Citation5 However, the results from us suggest that an additional mechanism acting through centromere misregulation also occurs in a significant proportion of human cancers where FBW7 is lost or Cyclin E1 is overexpressed.Citation6
Figure 1. A schematic model of FBW7 defects leading to Cyclin E1 overexpression, CENP-A Ser18 hyper-phosphorylation and chromosomal instability. In FBW7 wild-type cells, CENP-A is successfully deposited at centromeres in late mitosis and early G1 cell cycle phases in the absence of ectopic Cyclin E1/CDK2 activity. In contrast, FBW7 null cells accumulate Cyclin E1, and Cyclin E1 itself is frequently amplified in many cancers. Excessive Cyclin E1/CDK2 activity promotes aberrant CENP-A phosphorylation at the Ser18 residue, reduced CENP-A deposition at the centromere, lagging chromosomes and bridges in mitosis, and micronuclei (MN) formation associated with tumor progression.
The exact mechanism responsible for reduced interaction between HJURP and CENP-A upon CENP-A phosphorylation on Ser18 is currently unclear. Previous structural and molecular studies indicated direct interactions between the HJURP N-terminal region and the CENP-A histone fold domain.Citation28,Citation29 In our study, the failure of a phospho-mimetic CENP-A to be efficiently recruited to LacO arrays by LacI-HJURP, and decreased in vitro binding of HJURP in CENP-A S18D mutants, imply a reduced ability to form a competitive pre-nucleosomal complex or less competitive chromatin incorporation.Citation6 Perhaps CENP-A Ser18 phosphorylation perturbs the stable interaction between HJURP N terminal Scm3 domain and CENP-A histone fold domain. Additionally, it is possible that centromeric CENP-A phosphorylated at Ser18 reduces efficient HJURP recruitment for new CENP-A loading at endogenous centromeres.
Regardless, these results suggest that the CENP-A N terminal tail, and specifically levels of Ser18 phosphorylation, modulate proper centromeric CENP-A nucleosome assembly and centromere function. Previous domain replacement experiments using histone H3 N terminal tail suggested that CENP-A N terminal tail is not absolutely essential for cell viability.Citation30 However, the possibility that phosphorylation modulates centromere function in a cancer context could not be ruled out in those experiments. The results shown in our research are consistent with many findings in the field showing that the CENP-A N terminal tail regulates centromere functions in multiple speciesCitation31,Citation32, including proper CENP-B function in human cellsCitation30,Citation33, epigenetic stability of centromeres in fission yeastCitation34, CENP-A protein stability in budding yeast and DrosophilaCitation31,Citation32, and meiosis and organismal fertility in Arabidopsis.Citation35 Interestingly, ectopic overexpression of a CENP-A S18A mutation led to modest but statistically insignificant chromosome missegregation in HeLa cells where most CENP-A is phosphorylatedCitation24; while in our publication, CENP-A S18D or forced hyper-phosphorylation in DLD1 cells induces micronuclei.Citation6 The mechanistic details of this pathway await further investigation to better understand its role in carcinogenesis, and for future applications to therapeutic intervention.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
Acknowledgements
This research was partially supported by University Cancer Research Fund (UCRF) to QZ. The authors also acknowledge grant support from the National Institutes of Health (NIH) R01 GM119011 (to G.K), NIH (K99/R00 CA160351), Department of Defense Peer Reviewed Cancer Research Program Career Development Award (W81XWH-15-1-0599), The V Foundation Scholar Award, Kimmel Scholar Award, Mary Kay Foundation Award and Susan G. Komen Career Catalyst Award (to Q.Z.).
Additional information
Funding
References
- McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528-38. doi:10.1038/embor.2012.61. PMID:22595889
- Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501-14. doi:10.1038/embor.2012.55. PMID:22565320
- Wang Z, Inuzuka H, Zhong J, Wan L, Fukushima H, Sarkar FH, Wei W. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 2012;586:1409-18. doi:10.1016/j.febslet.2012.03.017. PMID:22673505
- Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297-300. doi:10.1038/45836. PMID:10499591
- Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, Petermann E. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013;32:3744-53. doi:10.1038/onc.2012.387. PMID:22945645
- Takada M, Zhang W, Suzuki A, Kuroda TS, Yu Z, Inuzuka H, Gao D, Wan L, Zhuang M, Hu L, et al. FBW7 Loss Promotes Chromosomal Instability and Tumorigenesis via Cyclin E1/CDK2-Mediated Phosphorylation of CENP-A. Cancer Res. 2017;77:4881-93. doi:10.1158/0008-5472.CAN-17-1240. PMID:28760857
- Westhorpe FG, Straight AF. Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol. 2013;25:334-40. doi:10.1016/j.ceb.2013.02.001. PMID:23490282
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923-37. doi:10.1038/nrg2466. PMID:19002142
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci U S A. 2000;97:1148-53. doi:10.1073/pnas.97.3.1148. PMID:10655499
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730-9. doi:10.1038/35087045. PMID:11483958
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209-26. doi:10.1083/jcb.153.6.1209. PMID:11402065
- Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol. 2003;160:25-39. doi:10.1083/jcb.200210005. PMID:12515822
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev cell. 2006;10:303-15. doi:10.1016/j.devcel.2006.01.014. PMID:16516834
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. The Journal of cell biology. 2011;194:229-43. doi:10.1083/jcb.201012017. PMID:21768289
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686-90. doi:10.1126/science.1206880. PMID:22053052
- Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7:e1002303. doi:10.1371/journal.pgen.1002303. PMID:21980305
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471-9. doi:10.1016/j.cell.2011.02.002. PMID:21335232
- Zhang W, Mao JH, Zhu W, Jain AK, Liu K, Brown JB, Karpen GH. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun. 2016;7:12619. doi:10.1038/ncomms12619. PMID:27577169
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511-6. PMID:12839935
- Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, Camps J, Ried T, Sung MH, Dalal Y. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenetics Chromatin. 2015;8:2. doi:10.1186/1756-8935-8-2. PMID:25788983
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, Kurumizaka H, Imhof A, Almouzni G. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell. 2014;53:631-44. doi:10.1016/j.molcel.2014.01.018. PMID:24530302
- Montes de Oca R, Gurard-Levin ZA, Berger F, Rehman H, Martel E, Corpet A, de Koning L, Vassias I, Wilson LO, Meseure D, et al. The histone chaperone HJURP is a new independent prognostic marker for luminal A breast carcinoma. Mol Oncol. 2015;9:657-74. doi:10.1016/j.molonc.2014.11.002. PMID:25497280
- Hu Z, Huang G, Sadanandam A, Gu S, Lenburg ME, Pai M, Bayani N, Blakely EA, Gray JW, Mao JH. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res: BCR. 2010;12:R18. doi:10.1186/bcr2487. PMID:20211017
- Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci U S A. 2013;110:11827-32. doi:10.1073/pnas.1300325110. PMID:23818633
- Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347-51. doi:10.1038/nature09323. PMID:20739937
- Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, et al. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell. 2015;32:68-81. doi:10.1016/j.devcel.2014.11.030. PMID:25556658
- Fachinetti D, Logsdon GA, Abdullah A, Selzer EB, Cleveland DW, Black BE. CENP-A Modifications on Ser68 and Lys124 Are Dispensable for Establishment, Maintenance, and Long-Term Function of Human Centromeres. Dev Cell. 2017;40:104-13. doi:10.1016/j.devcel.2016.12.014. PMID:28073008
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci U S A. 2010;107:1349-54. doi:10.1073/pnas.0913709107. PMID:20080577
- Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901-6. doi:10.1101/gad.2045111.
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1056-66. doi:10.1038/ncb2805. PMID:23873148
- Au WC, Dawson AR, Rawson DW, Taylor SB, Baker RE, Basrai MA. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics. 2013;194:513-8. doi:10.1534/genetics.113.149898. PMID:23525333
- Malik HS, Vermaak D, Henikoff S. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc Natl Acad Sci U S A. 2002;99:1449-54. doi:10.1073/pnas.032664299. PMID:11805302
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Developmental cell. 2015;33:314-27. doi:10.1016/j.devcel.2015.03.020. PMID:25942623
- Folco HD, Campbell CS, May KM, Espinoza CA, Oegema K, Hardwick KG, Grewal SI, Desai A. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr Biol. 2015;25:348-56. doi:10.1016/j.cub.2014.11.060. PMID:25619765
- Ravi M, Shibata F, Ramahi JS, Nagaki K, Chen C, Murata M, Chan SW. Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 2011;7:e1002121. doi:10.1371/journal.pgen.1002121. PMID:21695238
