ABSTRACT
Glioma remains one of the most aggressive and lethal cancers in central nervous system. Temozolomide (TMZ) is the most commonly used chemotherapeutic agent in gliomas. However, therapeutic benefits of TMZ could be very limited and all patients would finally suffer from tumor progression as the tumors develop resistance to TMZ. In this study, we aim to investigate the underlying mechanism of chemoresistance in glioma cell line and to identify whether there is still a close link between epithelial-mesenchymal transition (EMT) and TMZ resistance in gliomas. The real-time RT-PCR and Western blotting were used to measure the expression of EMT markers in TMZ-resistant cells. The migration and invasion assays were conducted to detect the cell motility activity in TMZ-resistant cells. The transfection was used to down-regulate the Cdc20 expression. The student t-test was applied for data analysis. We established stable TMZ-resistant glioma cells and designated as TR. Our results revealed that TR cells exhibited a significantly increased resistance to TMZ compared with their parental cells. Moreover, TMZ-resistant cells had acquired EMT-like changes. For the mechanism study, we measured a significant increased expression of CDC20 and decreased expression of Bim in TR cells. Moreover, upon suppression of CDC20 by shRNA transfection, TR cells underwent a reverse of EMT features. Importantly, knockdown of CDC20 enhanced the drug sensitivity of TR cells to TMZ. Our results suggested that inactivation of CDC20 could contribute to the future therapy that possibly overcomes drug resistance in human cancers.
Introduction
One of the leading causes of cancer death is brain cancer before age 40 years, which is characterized by high-grade proliferation and invasion [Citation1]. Cancers of the brain and other nervous system are the second most common cancer type account for 26% of all childhood cancers [Citation2]. Cerebral glioma is the most common brain tumor with high angiogenic malignancy [Citation3]. Being an aggressive tumour, glioma responds poorly to common therapeutic treatments including surgery, radiation, and conventional concomitant and adjuvant chemotherapy with temozolomide (TMZ). Although treatment options advanced during the past decades, the median survival of glioma is about 14.6 months and five-year survival is less than 10% [Citation4]. TMZ is an oral DNA-alkylating agent and the principal first-line chemotherapeutic agent used for treating patients with glioblastoma [Citation2,Citation5]. One study showed prediction of cell death responsiveness and sensitization of glioma cells to TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) and TMZ [Citation6]. However, therapeutic benefits of TMZ could be very limited and all patients would finally suffer from tumor progression as their tumors develop resistance to TMZ [Citation7]. Currently, the therapy is just palliative. One of the most popular events of TMZ-resistance is the expression of O6-methylguanine-DNA methyltransferase (MGMT) which defenses the mutagenic effects of alkylating agents on cellular genome [Citation8–10]. Thus, there is a need to characterize novel biological targets or signaling pathways that may responsible for tumor progression, in order to provide a better prognosis for patients with cancers [Citation11].
The molecular mechanism underlining TMZ resistance is still ambiguous [Citation12–14]. Accumulating evidence has proved that epithelial-mesenchymal transition (EMT) was closely related to chemoresistance [Citation15–20]. EMT was initially observed in embryonic development in which cells lost epithelial features and acquired mesenchymal characteristics in order to enhance motility and invasion [Citation21,Citation22]. Generally, EMT is accompanying with decreased expression of epithelial cell biomarkers (e.g. E-cadherin, Zo-1 and β-catenin) and increased expression of mesenchymal cell biomarkers (e.g. Vimentin, N-cadherin, Slug, Snail, and zinc finger E-box binding homeobox 1/2 (ZEB 1/2)) [Citation23–25]. Recent studies have shown that this process is also important in tumor progression and metastasis [Citation24,Citation26–31]. Yang et al. demonstrated that chronic oxaliplatin resistance induces EMT in colorectal cancer cell lines [Citation32]. Acquisition of EMT phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the Notch signaling pathway [Citation33]. Snail confers resistance to paclitaxel, adriamycin and radiotherapy by inhibiting p53-mediated apoptosis [Citation34]. However, recent studies in both breast cancer and pancreatic cancer have further demonstrated that acquisition of EMT is not required for invasion and metastasis, but contributes to chemoresistance [Citation35,Citation36]. And suppression of EMT contributes to enhanced sensitivity to gemcitabine treatment and increased overall survival of mice [Citation35]. Recently, it is also reported that blocking the EMT pathway abrogated resistance to anti-folate chemotherapy in lung cancer [Citation37]. In glioma cells, Siebzehnrubl and colleagues found that ZEB1 pathway regulated MGMT expression via miR-200c and c-MYB to promote initiation, invasion and chemoresistance [Citation38]. Furthermore, it is reported that in MGMT negative gliomas, activation of Wnt/β-catenin and Akt contribute to the EMT-like changes and TMZ resistance [Citation39].
In this study, we aim to investigate the underlying mechanism of chemoresistance in glioma cell line. We also aim to identify whether there is still a close link between EMT and TMZ resistance in gliomas, which will contribute to the future therapy that possibly overcome drug resistance. Relatively, TMZ-sensitive SNB19 and T98G glioma cell lines were treated with 100 μM TMZ for more than 6 months and the TMZ-resistant cells were established and designated as SNB19/TR and T98G/TR. We identified that the development of TMZ resistance in these two glioma cell lines was closely association with the acquisition of EMT-like changes, including both mesenchymal features change and the EMT biomarkers molecular alterations. We further found that the anaphase-promoting complex/cyclosome (APC/C)-associated protein CDC20 (cell division cycle 20 homolog), was activated in both TMZ-resistant cells. It is known that CDC20, an activator of the ligase APC/C, plays an oncogenic role in tumorigenesis via targeting its downstream substrates for ubiquitination and degradation [Citation40]. Bim, the downstream target of CDC20 [Citation41], was significantly decreased in these chemoresistant cells. These finding suggested that CDC20 pathway may contribute to the EMT and chemoresistance of SNB19/TR and T98G/TR cells.
Results
SNB19/TR and T98G/TR cells showed TMZ-resistance and morphologic changes
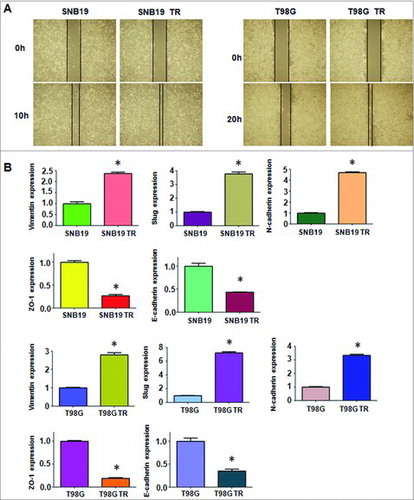
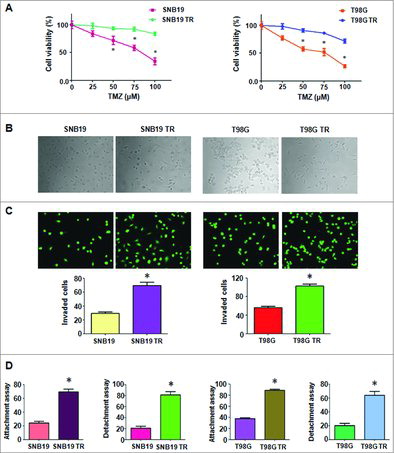
We established SNB19 and T98G TMZ-resistance cells with continuous stepwise selection in increasing concentrations of TMZ for more than 6 months. As shown in A, our MTT assay results suggest that SNB19/TR and T98G/TR were successfully established. Significantly different light-microscopic appearance was noted between TMZ-resistance glioma cells and their parental cells (B). SNB19/TR and T98G/TR cells decreased cell polarity and intercellular adhesion, increased formation of pseudopodia and intercellular separation (B), which displayed a typical mesenchymal phenotype.
Figure 1. TMZ-resistant cells exhibited EMT phenotype. A. MTT assay was conducted in parental and TMZ-resistant glioma cells. * P<0.05 vs TR cells. B. Cell morphology was observed by microscopy in parental and TMZ-resistant cells. C. Top panel: Invasion assay was performed to measure the invasive capacity in parental and TMZ-resistant glioma cells. Bottom panel: Quantitative results are illustrated for top panel. * P<0.05 vs control. D. Cell attachment and detachment assays were assessed in parental and TMZ-resistant cells. * P<0.05 vs control.
SNB19/TR and T98G/TR cells acquired EMT feature
EMT was described to associate with cells aggressive characteristics, such as cell attachment, detachment, migration, and invasion. Consistent with this, our data provided that TMZ-resistant glioma cells exhibited significantly increased invaded cells compared with parental cells by Transwell assay (C). We also found that TMZ-resistant cells displayed enhanced capacity of attachment and detachment (D). Moreover, we observed that TMZ-resistant cells exhibited enhanced motility activity by wound healing assay (A). Taken together, our findings revealed that TMZ-resistant cells acquired EMT feature.
SNB19/TR and T98G/TR cells went through EMT molecular changes
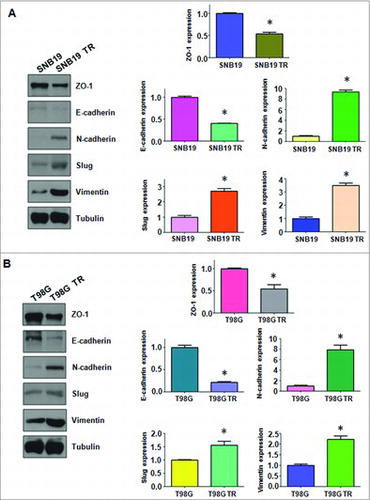
Multiple biological changes of TMZ-resistant cells were determined. Several important EMT molecular markers were detected in TMZ-resistant cells and their paired parental cells. As shown in B, q-PCR analysis showed the mRNA levels of mesenchymal molecule such as Slug, Vimentin, and N-cadherin, were significantly up-regulated in TMZ-resistant cells. While, the mRNA levels of epithelial markers Zo-1 and E-cadherin were significantly decreased in resistant cells. In line with this concept, similar EMT markers changes at protein levels in TMZ-resistant cells were determined by Western blotting analysis (A & B).
Figure 3. TMZ-resistant cells hade EMT marker changes. A. Left panel: Western blotting analysis was used to detect the expression of ZO-1, E-cadherin, N-cadherin, Slug, and Vimentin in SNB19 and SNB19/TR cells. Right panel: Quantitative results are illustrated for left panel. * P<0.05 vs control. B. Left panel: Western blotting analysis was performed to measure the expression of ZO-1, E-cadherin, N-cadherin, Slug, and Vimentin in T98G and T98G/TR cells. Right Panel: Quantitative results are illustrated for left panel. * P<0.05 vs control.
Over-expression of CDC20 was associated with TMZ-resistance in SNB19/TR and T98G/TR
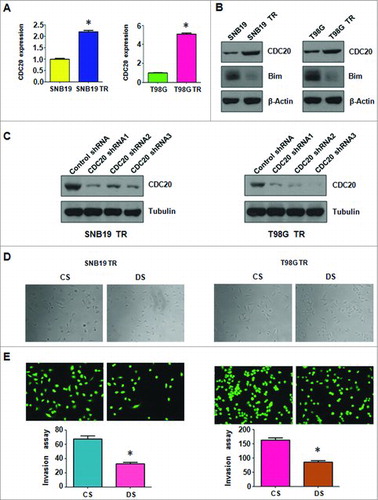
As CDC20 has been involved in EMT in many cancers, we detected the expression of CDC20 in TMZ-resistant glioma cells. We proved that both mRNA and protein levels of CDC20 were markedly increased in SNB19/TR and T98G/TR cell lines (A). Moreover, Bim, one of the downstream targets of CDC20 was significantly decreased in TMZ-resistant glioma cells (A). These findings suggested that CDC20 is associated with TMZ induced resistance in glioma cells.
Figure 4. TMZ-resistant cells had high expression of CDC20. A. Real-time RT-PCR assay was performed to detect the expression of CDC20 in parental and TMZ-resistant cells. * P<0.05 vs control. B. Western blotting analysis was performed to detect the expression of CDC20 in parental and TMZ-resistant cells. C. Western blotting analysis was performed to detect the efficacy of CDC20 shRNA transfection. D. Cell morphology was taken by microscopy in TMZ-resistant cells transfected with CDC20 shRNA. CS: Control shRNA; DS: CDC20 shRNA. E. Invasion assay were performed in TMZ-resistant cells transfected with CDC20 shRNA. CS: Control shRNA; DS: CDC20 shRNA.
Inhibition of CDC20 reversed EMT features in SNB19/TR and T98G/TR
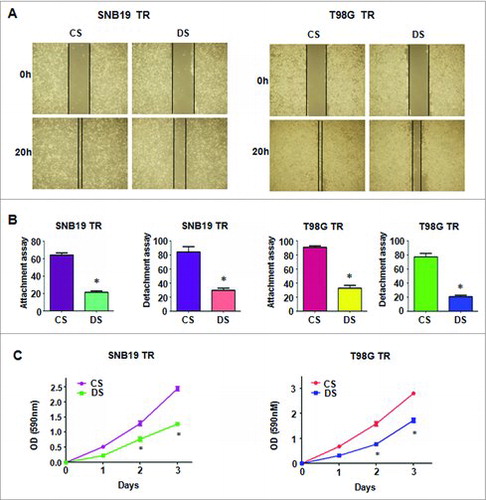
Significant inhibition of CDC20 expression was detected in SNB19/TR and T98G/TR cells after transfected with three shRNAs which targeting CDC20 sequence (B). ShRNA3 was used for the subsequent transfection. Suppressed expression of CDC20 partially reversed the EMT morphological features to mesenchymal–epithelial transition (MET) in TMZ-resistant glioma cells (C). Transwell assay further determined that depletion of CDC20 could impair the invasion capacity of SNB19/TR and T98G/TR cells (D). The motility of TMZ-resistant cells was further determined by Wound healing assay. As shown in A, the migration of both SNB19/TR and T98G/TR cells was slow down by CDC20 shRNA transfection. Knock down of CDC20 could also abrogate the attachment and detachment capacity of TMZ-resistant glioma cells (B). These results showed that inhibition of CDC20 could reverse the EMT characteristics of TMZ-resistant glioma cells.
Figure 5. Depletion of CDC20 inhibited motility and enhanced TMZ sensitivity in TMZ-resistant cells. A. Wound healing assays were used to detect the motility in TMZ-resistant cells transfected with CDC20 shRNA B. Cell attachment and detachment assays were measured in TMZ-resistant cells transfected with CDC20 shRNA. * P<0.05 vs control. CS: control shRNA; DS: CDC20 shRNA. C. MTT assay was performed in TMZ-resistant cells treated with CDC20 shRNA. CS: control shRNA; DS: CDC20 shRNA. *, P<0.05 compared with control shRNA.
Suppression of CDC20 enhanced the sensitivity of TMZ-resistant glioma cells to TMZ treatment
We further determine whether suppression of CDC20 could affect the drug sensitivity of the TMZ-resistant cells. MTT assay results showed that TMZ-mediated cell growth inhibition in the resistant cells was partially enhanced by CDC20 shRNA transfection (C). This data revealed that CDC20 depleted glioma cells could be more sensitive to TMZ treatment, and CDC20 could be a promising target to overcome drug resistance in human cancers.
Suppression of CDC20 regulated the expression of EMT markers
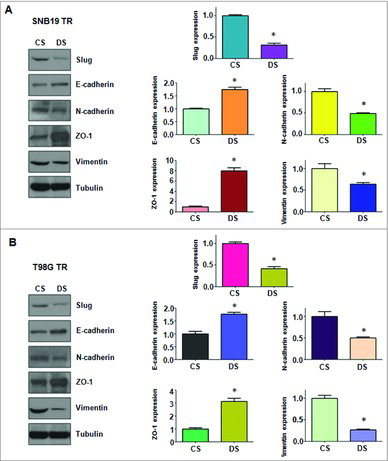
After transfection with shRNA in SNB19/TR and T98G/TR cells, the protein levels of EMT markers were measured to further determine the effects of CDC20 suppression. As shown in , higher levels of epithelial marker E-cadherin and Zo-1 were promoted in TMZ-resistant glioma cells after CDC20 depletion, whereas the expression of mesenchymal markers including N-cadherin, Vimentin, and Slug was significantly down-regulated in TMZ-resistant cells with CDC20 shRNA transfection. Taken together, these findings suggested that CDC20 is closely associated with the modulation of EMT progress in TMZ-resistant glioma cells.
Figure 6. Depletion of CDC20 regulated protein levels of EMT markers in TMZ-resistant cells. A. Left panel: Western blotting analysis was used to detect the expression of EMT markers in SNB19/TR cells after CDC20 depletion. Right panel: Quantitative results are illustrated for left panel. * P<0.05 vs control. B. Left panel: Western blotting analysis was performed to measure the expression of EMT markers in T98G/TR cells after CDC20 depletion. Right Panel: Quantitative results are illustrated for left panel. * P<0.05 vs control.
Discussion
Although new treatment advances, glioma is associated with a poor overall survival [Citation4]. Concomitant and adjuvant chemotherapy with TMZ is used in the current standard-of-care for patients with newly diagnosed glioma. However, TMZ-resistance development in glioma patients becomes a major obstacle which is responsible for the therapy failure. Expect the common known mechanism of TMZ-resistance mediated by MGMT, there are also other mechanisms exist in chemoresistance of gliomas [Citation42–44]. EMT is currently undergoing extensive research in tumor progression, metastasis and chemoresistance.
In this study, we established stable TMZ-resistant SNB19 and T98G cell lines, and such drug-resistance was kept upon the removal of TMZ for a considerable time interval. Our results revealed that SNB19/TR and T98G/TR cells exhibited a significantly increased resistance to TMZ compared with their parental cells. Analysis of SNB19/TR and T98G/TR cells could be of great importance to explore the new mechanism of TMZ-resistance in gliomas. Recent researches have demonstrated that several chemotherapeutic agent-resistant tumor cell lines exhibited EMT-like changes, consisting of both morphology and molecular markers [Citation32,Citation33,Citation37–39]. It was also reported that treatment with frizzled-related protein 4 (sFRP4), a Wnt/ß-catenin antagonist, could help to improve response to TMZ chemotherapeutics in gliomas by the reversal of EMT [Citation45]. In this series of experiments, we have observed the mesenchymal-like phenotypic changes in both SNB19/TR and T98G/TR cells. Also we have found that the TMZ-resistant cells underwent EMT biomarkers alternations, including significantly decreased expression of epithelial adhesion molecules E-cadherin and Zo-1, and a concurrent prominent increased expression of mesenchymal markers (vimentin and N-cadherin) and EMT-related transcription factors Slug. Moreover, we also found that TMZ-resistant cells had acquired an increased invasive and migrate capacity. These findings suggested that EMT could play an important role in the development of TMZ resistance in glioma cells.
We further explore new cell signaling moleculars that might be involved in the regulation of EMT and TMZ resistance in glioma cells. We detected for the first time that CDC20 pathway was activated in SNB19/TR and T98G/TR cells compared with their parental cells. We also found the expression of Bim, which was a downstream target of CDC20, decreased in the resistant cells. Recently, it was reported that competitive binding between Id1 and E2F1 to CDC20 regulated E2F1 degradation and thymidylate synthase expression to promote esophageal cancer fluorouracil (5-FU)-resistance [Citation46]. An RNA-seq revealed CDC20 might be related to irinotecan resistance in colorectal cancer cell lines [Citation47]. A new coumarin substituted hydrazide-hydrazone derivatives exhibited potent anticancer property to overcome drug resistance in pancreatic carcinoma cells at least partially by down-regulation of CDC20 [Citation48]. Consistently, human adult T cell leukemia cells that acquire elevated CDC20 activity via expressing the Tax viral oncoprotein exhibit reduced Bim levels and resistance to anticancer agents [Citation41]. And upon knockdown, CDC20 and multiple APC-core components were identified to sensitize resistant cancer cells to chemoradiation in a Bim-dependent manner [Citation41]. In line with this, we found an activated CDC20 and decreased Bim in TMZ-resistant glioma cells. We also proved that suppression of CDC20 by shRNA transfection enhanced the drug sensitivity of SNB19/TR and T98G/TR cells. One study showed that Cdc20 could control cell invasion via regulation of transcription factor SOX2 in glioblastoma cells [Citation49]. Therefore, TMZ-resistant cells enhanced migration and invasion partly due to upregulation of SOX2. It is worth noting that CDC20 depletion had also caused a significant reverse of TMZ-resistant glioma cells from EMT to MET features. Although the exact mechanism of CDC20-regulated EMT and drug resistance in cancer cells needs to be revealed in future researches, targeting CDC20 could be a promising option to overcome chemoresistance of human cancers.
Materials and methods
Reagents and antibodies
Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased from Gibco (MGC803; Grand Island, NY). Penicillin/streptomycin was obtained from HyClone™ (Logan, Utah, USA). TMZ (CAS number phr1437), DMSO (Dimethyl sulfoxide, CAS number d8779) and MTT (3-4,5-dimethyl-2- thiazolyl-2, 5-diphenyl-2-H-tetrazolium bromide, CAS number 57360-69-7) were purchased from Sigma-Aldrich (St. Louis, MO, USA). TMZ was diluted in DMSO and stocked at -20˚C. TRIzol, Lipofectamine 2000, and plus reagents were obtained from Invitrogen (Carlsbad, CA). Penicillin/streptomycin, RevertAid First Strand cDNA Synthesis Kit, and SYBR® Select Master mix were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Primary antibodies for E-cadherin (#3195, 1:1000), N-cadherin (#13116, 1:1000), vimentin (#5741, 1:1000), Zo-1 (#8193, 1:1000) and Slug (#9585, 1:1000) were purchased from Cell Signaling Technology (Danvers, MA, USA). All secondary antibodies were purchased from Thermo Fisher Scientific. Monoclonal anti-Tubulin (T9028, 1:5000) was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell culture
The SNB19 and T98G human glioma cell lines were purchased from the Chinese Academy of Sciences (Shanghai, China) and maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37˚C with 5% CO2. As we have identified the half-maximal inhibitory concentration (IC50) of TMZ for SNB19 and T98G were 75 μM to 100 μM, we therefore selected 100 μM TMZ to prepare for TMZ-resistant glioma cells. After over 6 months’ treatment of 100 μM TMZ, TMZ-resistant cells were established and designated as SNB19/TR and T98G/TR. The IC50 was measured at monthly intervals to make sure the stable resistance to TMZ during the period of culturing in drug-free medium.
MTT assays
SNB19, T98G and their TMZ-resistant cells (2.5 × 103 cells/well) were seeded in 96-well plates and cultured overnight. Different concentrations of TMZ were added to the medium for 48 h. Then MTT assays were performed to determine cell viability following the manufacturer's protocols. Briefly, added 10 μl MTT solutions (0.5 mg/ml) to each well and incubated for 4 h. Removed the liquid supernatant and added 100 μl DMSO to each well. We used Multimode Reader of SpectraMax M5 (Molecular Devices, Sunnyvale, CA, USA) to determine the absorbance of each well at 490 nm. Independent experiments were repeated in triplicate.
Morphological analysis
SNB19, T98G and their TMZ-resistant cells were grown to about 70% confluence and digital images were captured with a Leica light microscope (DMI3000 B, Germany). These cell lines for Morphologic characteristics consistent with EMT were compared with their parental cells.
Transwell assay
SNB19, T98G and their TMZ-resistant cells were suspended with 200 μl serum-free DMEM at a density of 1.0 × 105 cells/well and seeded in the upper-chamber of the Matrigel precoated Transwell inserts (24-well insert; Corning Incorporated, Corning, NY, USA). 500 μL of complete medium containing 10% FBS was added in the lower-chamber. About 24 h later, the cells on the upper membrane surface were removed using a cotton bud. The migratory cells on lower membrane surface were stained with Calcein-AM, photographed under a microscope and counted using a microplate reader.
Cell attachment and detachment assay
Cell attachment and detachment abilities of the above four cell lines were determined. For attachment assay, seeded 5 × 104 cells per well in 24-well plates and incubated for 1 h. Then, removed the unattached cells and counted attached cells. For cell detachment assay, seeded 5 × 104 cells in 24-well plates and incubated for 24 h. The cells were treated with 0.05% trypsin for 3 min and the detached cells were counted. Data were presented as a percentage of the attached or detached cells to total cells.
Wound healing assay
Glioma cells were seeded in six-well plate and grew to 90% confluence. A rectangular wound on monolayers was generated by a sterile 100 μl pipette tip. Photographic images were taken at the lesion border at the designated time course using an inverted microscope (Olympus, IX71).
Quantitative real-time reverse transcription-PCR (Q-PCR) analysis
Total RNAs were extracted with Trizol reagent according to the manufacturer's introduction. The relative quantitative real-time PCR reactions were performed using Power SYBR Green PCR Master Mix [Citation50]. The q-PCR results were calculated by 2−▹▹Ct method. The primers used in PCR reaction are listed below: E-cadherin, forward primer (5’-GAA GTG TCC GAG GAC TTT GG-3’) and reverse primer (5’-CAG TGT CTC TCC AAA TCC GAT A-3’); Vimentin, forward primer (5’-TGT CCA AAT CGA TGT GGA TGT TTC-3’) and reverse primer (5’-TTG TAC CAT TCT TCT GCC TCC TG-3’); Snail, forward primer (5’-CGG AAG CCT AAC TAC AGC GA-3’) and reverse primer (5’-GGA CAG AGT CCC AGA TGA GC-3’); Slug, forward primer (5’-CAT GCC TGT CAT ACC ACA AC-3’) and reverse primer (5’-GGT GTC AGA TGG AGG AGG G-3’); N-cadherin, forward primer (5’-CCT GCG CGT GAA GGT TTG CC- 3’) and reverse primer (5’-CCA AGC CCC GCA CCC ACA AT-3’); GAPDH, forward primer (5’-CAG CCT CAA GAT CAT CAG CA-3’) and reverse primer (5’-TGT GGT CAT GAG TCC TTC CA-3’).
Transfection
Short hairpin RNAs (shRNAs) of CDC20 were transfected to glioma cells, using the Lipofectamine™ 2000 transfection reagent by following the manufacturer's instruments [Citation51]. The shRNA sequences that targeting human CDC20 were listed below. Sense: 5′-GGA GAA UUA UGG GUG UCA ATT -3′; antisense 5′-UUG ACA CCC AUA AUU CUC CTT -3′.
Western blotting analysis
Whole-cell protein was isolated using cell lysis buffer supplemented with phenylmethylsulfonyl fluoride (PMSF) (Solarbio Inc, Beijing, China). Protein concentrations were quantified by BCA assay (Solarbio Inc, Beijing, China). Boiled the isolated protein with 1 × Loading buffer and separated the denatured protein samples by SDS-PAGE on a 10% polyacrylamide gel. The samples were transferred to a polyvinylidene difluoride membrane (PVDF) (Millipore Corp., Billerica, MA, USA). After blocked with 5% nonfat milk for 1 h, the membranes were probed with the appropriate primary antibody overnight at 4 °C. The next morning, the membranes were washed with TBST and incubated with appropriate second antibody for around an hour at room temperature. The protein bands were visualized by electrochemiluminescence (ECL) assay.
Statistical analysis
GraphPad Prism 4.0 (Graph Pad Software, La Jolla, CA) was conducted with all of the data. The ANOVA was applied for data analysis. Statistical analyses were expressed as mean ± SD (standard deviations) of triplicates determinants. p<0.05 was considered statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Science. 2009;100:2235–41. doi:10.1111/j.1349-7006.2009.01308.x. PMID:19737147.
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: A cancer. Journal for Clinicians. 2017;67:7–30. doi:10.3322/caac.21387.
- Aibaidula A, Lu JF, Wu JS, Zou HJ, Chen H, Wang YQ, Qin ZY, Yao Y, Gong Y, Che XM, et al. Establishment and maintenance of a standardized glioma tissue bank: Huashan experience. Cell and Tissue Banking. 2015;16:271–81. doi:10.1007/s10561-014-9459-4. PMID:24929994.
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncology. 2009;10:459–66. doi:10.1016/S1470-2045(09)70025-7. PMID:19269895.
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. New England Journal of Medicine. 2005;352:997–1003. doi:10.1056/NEJMoa043331. PMID:15758010.
- Weyhenmeyer BC, Noonan J, Wurstle ML, Lincoln FA, Johnston G, Rehm M, Murphy BM. Predicting the cell death responsiveness and sensitization of glioma cells to TRAIL and temozolomide. Oncotarget. 2016;7:61295–311. doi:10.18632/oncotarget.10973. PMID:27494880.
- Nagel ZD, Kitange GJ, Gupta SK, Joughin BA, Chaim IA, Mazzucato P, Lauffenburger DA, Sarkaria JN, Samson LD. DNA Repair Capacity in Multiple Pathways Predicts Chemoresistance in Glioblastoma Multiforme. Cancer Research. 2017;77:198–206. doi:10.1158/0008-5472.CAN-16-1151. PMID:27793847.
- Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M, Wick W. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–8. doi:10.1158/1078-0432.CCR-07-1719. PMID:18483356.
- Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clinical Cancer Research. 2000;6:2585–97. PMID:10914698.
- Wickstrom M, Dyberg C, Milosevic J, Einvik C, Calero R, Sveinbjornsson B, Sanden E, Darabi A, Siesjo P, Kool M, et al. Wnt/beta-catenin pathway regulates MGMT gene expression in cancer and inhibition of Wnt signalling prevents chemoresistance. Nature Communications. 2015;6:8904. doi:10.1038/ncomms9904. PMID:26603103.
- Huse JT, Holland EC. Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nature Reviews Cancer. 2010;10:319–31. doi:10.1038/nrc2818. PMID:20414201.
- Rocha CR, Kajitani GS, Quinet A, Fortunato RS, Menck CF. NRF2 and glutathione are key resistance mediators to temozolomide in glioma and melanoma cells. Oncotarget. 2016;7:48081–92. doi:10.18632/oncotarget.10129. PMID:27344172.
- Li H, Yuan X, Yan D, Li D, Guan F, Dong Y, Wang H, Liu X, Yang B. Long Non-Coding RNA MALAT1 Decreases the Sensitivity of Resistant Glioblastoma Cell Lines to Temozolomide. Cell Physiol Biochem. 2017;42:1192–201. doi:10.1159/000478917. PMID:28668966.
- Yu Z, Zhao G, Xie G, Zhao L, Chen Y, Yu H, Zhang Z, Li C, Li Y. Metformin and temozolomide act synergistically to inhibit growth of glioma cells and glioma stem cells in vitro and in vivo. Oncotarget. 2015;6:32930–43. doi:10.18632/oncotarget.5405. PMID:26431379.
- Zheng HC. The molecular mechanisms of chemoresistance in cancers. Oncotarget. 2017;8(35):59950–59964. doi:10.18632/oncotarget.19048. PMID: 28938696.
- Lu C, Shervington A. Chemoresistance in gliomas. Molecular and Cellular Biochemistry. 2008;312:71–80. doi:10.1007/s11010-008-9722-8. PMID:18259841.
- Ahmed N, Abubaker K, Findlay J, Quinn M. Epithelial mesenchymal transition and cancer stem cell-like phenotypes facilitate chemoresistance in recurrent ovarian cancer. Current Cancer Drug Targets. 2010;10:268–78. doi:10.2174/156800910791190175. PMID:20370691.
- Liao H, Bai Y, Qiu S, Zheng L, Huang L, Liu T, Wang X, Liu Y, Xu N, Yan X, et al. MiR-203 downregulation is responsible for chemoresistance in human glioblastoma by promoting epithelial-mesenchymal transition via SNAI2. Oncotarget. 2015;6:8914–28. doi:10.18632/oncotarget.3563. PMID:25871397.
- Zhang Y, Zeng S, Ma J, Deng G, Qu Y, Guo C, Shen H. Nestin overexpression in hepatocellular carcinoma associates with epithelial-mesenchymal transition and chemoresistance. J Exp Clin Cancer Res. 2016;35:111. doi:10.1186/s13046-016-0387-y. PMID:27412382.
- Zhou P, Li B, Liu F, Zhang M, Wang Q, Liu Y, Yao Y, Li D. The epithelial to mesenchymal transition (EMT) and cancer stem cells: Implication for treatment resistance in pancreatic cancer. Molecular Cancer. 2017;16:52. doi:10.1186/s12943-017-0624-9. PMID:28245823.
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature Reviews Molecular Cell Biology. 2014;15:178–96. doi:10.1038/nrm3758. PMID:24556840.
- Kong D, Li Y, Wang Z, Sarkar FH. Cancer Stem Cells and Epithelial-to-Mesenchymal Transition (EMT)-Phenotypic Cells: Are They Cousins or Twins? Cancers. 2011;3:716–29. doi:10.3390/cancers30100716. PMID:21643534.
- Ksiazkiewicz M, Markiewicz A, Zaczek AJ. Epithelial-mesenchymal transition: A hallmark in metastasis formation linking circulating tumor cells and cancer stem cells. Pathobiology. 2012;79:195–208. doi:10.1159/000337106. PMID:22488297.
- Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. Journal of Clinical Investigation. 2009;119:1429–37. doi:10.1172/JCI36183.
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nature Reviews Molecular Cell Biology. 2006;7:131–42. doi:10.1038/nrm1835. PMID:16493418.
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nature Reviews Cancer. 2009;9:265–73. doi:10.1038/nrc2620. PMID:19262571.
- Samatov TR, Tonevitsky AG, Schumacher U. Epithelial-mesenchymal transition: Focus on metastatic cascade, alternative splicing, non-coding RNAs and modulating compounds. Molecular Cancer. 2013;12:107. doi:10.1186/1476-4598-12-107. PMID:24053443.
- Yang S, Liu Y, Li MY, Ng CSH, Yang SL, Wang S, Zou C, Dong Y, Du J, Long X, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/beta-catenin signaling pathway and EMT in non-small cell lung cancer. Molecular Cancer. 2017;16:124. doi:10.1186/s12943-017-0700-1. PMID:28716029.
- Tran HD, Luitel K, Kim M, Zhang K, Longmore GD, Tran DD. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Research. 2014;74:6330–40. doi:10.1158/0008-5472.CAN-14-0923. PMID:25164016.
- Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, Schafer R, van Diest P, Voest E, van Oudenaarden A, et al. Plasticity between Epithelial and Mesenchymal States Unlinks EMT from Metastasis-Enhancing Stem Cell Capacity. Cell Reports. 2016;14:2281–8. doi:10.1016/j.celrep.2016.02.034. PMID:26947068.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi:10.1016/j.cell.2011.02.013. PMID:21376230.
- Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R, Gray MJ, Cheng H, Hoff PM, Ellis LM. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147–53. doi:10.1158/1078-0432.CCR-06-0038. PMID:16857785.
- Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, Ali S, Abbruzzese JL, Gallick GE, Sarkar FH. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Research. 2009;69:2400–7. doi:10.1158/0008-5472.CAN-08-4312. PMID:19276344.
- Kajiyama H, Shibata K, Terauchi M, Yamashita M, Ino K, Nawa A, Kikkawa F. Chemoresistance to paclitaxel induces epithelial-mesenchymal transition and enhances metastatic potential for epithelial ovarian carcinoma cells. International Journal of Oncology. 2007;31:277–83. PMID:17611683.
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–30. doi:10.1038/nature16064. PMID:26560028.
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–6. doi:10.1038/nature15748. PMID:26560033.
- Liang SQ, Marti TM, Dorn P, Froment L, Hall SR, Berezowska S, Kocher G, Schmid RA, Peng RW. Blocking the epithelial-to-mesenchymal transition pathway abrogates resistance to anti-folate chemotherapy in lung cancer. Cell Death & Disease. 2015;6:e1824. doi:10.1038/cddis.2015.195.
- Siebzehnrubl FA, Silver DJ, Tugertimur B, Deleyrolle LP, Siebzehnrubl D, Sarkisian MR, Devers KG, Yachnis AT, Kupper MD, Neal D, et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Molecular Medicine. 2013;5:1196–212. doi:10.1002/emmm.201302827. PMID:23818228.
- Yi GZ, Liu YW, Xiang W, Wang H, Chen ZY, Xie SD, Qi ST. Akt and beta-catenin contribute to TMZ resistance and EMT of MGMT negative malignant glioma cell line. Journal of the Neurological Sciences. 2016;367:101–6. doi:10.1016/j.jns.2016.05.054. PMID:27423571.
- Wang L, Zhang J, Wan L, Zhou X, Wang Z, Wei W. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol Ther. 2015;151:141–51. doi:10.1016/j.pharmthera.2015.04.002. PMID:25850036.
- Wan L, Tan M, Yang J, Inuzuka H, Dai X, Wu T, Liu J, Shaik S, Chen G, Deng J, et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Developmental Cell. 2014;29:377–91. doi:10.1016/j.devcel.2014.04.022. PMID:24871945.
- Fodale V, Pierobon M, Liotta L, Petricoin E. Mechanism of cell adaptation: when and how do cancer cells develop chemoresistance? Cancer Journal. 2011;17:89–95. doi:10.1097/PPO.0b013e318212dd3d.
- Goellner EM, Grimme B, Brown AR, Lin YC, Wang XH, Sugrue KF, Mitchell L, Trivedi RN, Tang JB, Sobol RW. Overcoming temozolomide resistance in glioblastoma via dual inhibition of NAD+ biosynthesis and base excision repair. Cancer Research. 2011;71:2308–17. doi:10.1158/0008-5472.CAN-10-3213. PMID:21406402.
- Munoz JL, Rodriguez-Cruz V, Greco SJ, Nagula V, Scotto KW, Rameshwar P. Temozolomide induces the production of epidermal growth factor to regulate MDR1 expression in glioblastoma cells. Mol Cancer Ther. 2014;13:2399–411. doi:10.1158/1535-7163.MCT-14-0011. PMID:25053824.
- Bhuvanalakshmi G, Arfuso F, Millward M, Dharmarajan A, Warrier S. Secreted frizzled-related protein 4 inhibits glioma stem-like cells by reversing epithelial to mesenchymal transition, inducing apoptosis and decreasing cancer stem cell properties. PloS One. 2015;10:e0127517. doi:10.1371/journal.pone.0127517. PMID:26030909.
- Li B, Xu WW, Guan XY, Qin YR, Law S, Lee NP, Chan KT, Tam PY, Li YY, Chan KW, et al. Competitive Binding Between Id1 and E2F1 to Cdc20 Regulates E2F1 Degradation and Thymidylate Synthase Expression to Promote Esophageal Cancer Chemoresistance. Clin Cancer Res. 2016;22:1243–55. doi:10.1158/1078-0432.CCR-15-1196. PMID:26475334.
- Li XX, Zheng HT, Peng JJ, Huang LY, Shi DB, Liang L, Cai SJ. RNA-seq reveals determinants for irinotecan sensitivity/resistance in colorectal cancer cell lines. Int J Clin Exp Pathol. 2014;7:2729–36. PMID:24966994.
- Nasr T, Bondock S, Youns M. Anticancer activity of new coumarin substituted hydrazide-hydrazone derivatives. Eur J Med Chem. 2014;76:539–48. doi:10.1016/j.ejmech.2014.02.026. PMID:24607878.
- Mao DD, Gujar AD, Mahlokozera T, Chen I, Pan Y, Luo J, Brost T, Thompson EA, Turski A, Leuthardt EC, et al. A CDC20-APC/SOX2 Signaling Axis Regulates Human Glioblastoma Stem-like Cells. Cell Reports. 2015;11:1809–21. doi:10.1016/j.celrep.2015.05.027. PMID:26074073.
- Li S, Zhang X, Zhang R, Liang Z, Liao W, Du Z, Gao C, Liu F, Fan Y, Hong H. Hippo pathway contributes to cisplatin resistant-induced EMT in nasopharyngeal carcinoma cells. Cell Cycle. 2017;16:1601–10. doi:10.1080/15384101.2017.1356508. PMID:28749195.
- Wang L, Hou Y, Yin X, Su J, Zhao Z, Ye X, Zhou X, Zhou L, Wang Z. Rottlerin inhibits cell growth and invasion via down-regulation of Cdc20 in glioma cells. Oncotarget. 2016;7:69770–82. PMID:27626499.