Biological oscillators have fascinated researchers for decades, and debates concerning the control systems that underlie them have often been fierce. One oscillator essential to all eukaryotes drives progression through the sequential events of the cell cycle. In the 1980s, genetic screens of yeast and bulk biochemical approaches using eggs and early embryos from various aquatic organisms uncovered the first known core cell cycle regulators, namely cyclin-dependent kinase (CDK) and cyclin. From these discoveries, an early controversy arose concerning the view of how cell cycle passage is mediated.Citation1 A perspective gathered from yeast studies was that completion of cell cycle events was required for progression to subsequent steps (like “dominoes”). Alternatively, work using early embryos of clam, starfish, and Xenopus suggested that the cell cycle oscillator functions by pushing forward independently without pause (like a “clock”), even in parthenogenetically-activated eggs or enucleated embryos. As CDK and cyclin homologues were found capable of serving as functional replacements between evolutionarily diverse organisms, it was indisputable that their roles had been preserved, while the layers of regulation upon the oscillator itself diverged to suit the respective organismic and developmental contexts in which it functions. This early debate revealed the importance of considering the context of the biochemical and molecular solutions that evolved.
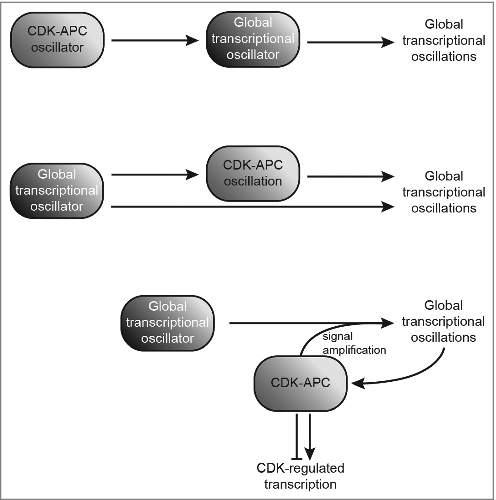
More recently, another debate has emerged concerning the nature of the central oscillator controlling periodic transcription in budding yeast. Textbook models suggest that an autonomous CDK-APC biochemical oscillator drives transcriptional oscillations (, top). However, the discovery that cell-cycle transcription factors interact in a network gave rise to a model that gene regulatory networks could drive the transcriptional program independent of CDK (, center).Citation2 Tests of these models have aimed to block oscillations in CDK activity and cell-cycle progression by holding cells at either low CDK activityCitation3,4 or high CDK activityCitation5,6, followed by examining the effects on cell cycle transcription. Surprisingly, blocks at either low or high CDK activity failed to impede certain transcriptional oscillations, and demonstrated that much of the periodic transcriptional program could be uncoupled from cell-cycle progression.Citation3,4,5 However, a recent publication suggested that merely minimal-but-sufficient cyclin/CDK activity—possibly by residual cyclin—led to the observations of autonomous transcriptional oscillations in the earlier published work.Citation7 The group utilized precisely regulated expression of cyclins, and implemented RNA-seq analysis and single-cell live imaging to reach the opposite conclusion: the CDK-APC oscillator is indeed the master regulator that drives periodic and global cell-cycle transcription.
Figure 1. Models past and present of the drivers of global transcriptional oscillations during the eukaryotic cell cycle in yeast. The CDK-APC oscillator controls a global transcriptional oscillator (top); a global transcriptional oscillator directly regulates oscillations in global transcription and CDK1-cyclin B activity, while CDK1-APC oscillations also impact global transcriptional oscillations (center); or, a global transcriptional oscillator drives periodic transcription and is influenced by signal amplification through CDK activity, and vice versa (bottom). All enclosed components indicated are control elements; components indicated by arrowheads are outputs.
To address this contradiction directly, Cho et al. analyzed data published by both groups.Citation6 Even with distinctions in experimental approach and variations in the engineered yeast strains, the two groups provide remarkably reproducible and complementary data. While it is tempting to speculate that subtle variations appeared due to differences in strains, experimental protocol, or quantitative analysis, Cho et al. provide compelling explanations for variations and provide a new set of experimental evidence underscoring a model in which CDKs are part of a transcriptional oscillator, but that periodic input from CDK is not required to drive global transcriptional oscillations. This is an important distinction that does not diminish the role of CDK-APC in impacting certain transcriptional programs. For example, SBF and Swi5/Ace2-regulated genes are inhibited by Clb2/CDK, so it certainly can have profound effects on global transcriptional oscillations.
Decreased amplitudes of global transcriptional oscillations are observed in both Cho et al. and Rahi et al. when CDK is locked in an off-state.Citation6,7 This observation raises a key question: what qualifies a behavior as “oscillatory/periodic”? What if experimental perturbation of a variable (or more than one!) merely leads to a significantly damped oscillation? The current study further highlights the possibility that CDK plays a critical role as signal amplifier to global transcriptional oscillations—posing a third and even more likely model (, bottom)—where CDK/APC collaborates with transcription factors to produce a robust transcriptional program during the cell cycle.
In summary, it should come as no surprise that evolution has provided fantastic solutions to ensure that eukaryotic proliferation is robust and highly adaptable to accommodate various organismic and developmental contexts. The study by Cho et al. further exemplifies the need to dig deeper into the fundamental—and sometimes complex—mechanisms that establish and drive eukaryotic cell cycle progression, with full acknowledgement that oscillator functions are influenced by cellular context and likely controlled by multiple layers of transcriptional and post-transcriptional regulation. And as flexibilities in how these systems interoperate likely exist, it is apparent—like the groups working to uncover these mysteries—that they work better together.
References
- Murray A, Kirschner, M. Dominoes and clocks: the union of two views of the cell cycle. Science. 1989;246(4930):614–21. doi:10.1126/science.2683077. PMID:2683077.
- Simon I, Barnett J, Hannett N, Harbison C, Rinaldi N, Volkert T, Wyrick J, Zeitlinger J, Gifford D, Jaakkola, T et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106(6):697–708. doi:10.1016/S0092-8674(01)00494-9. PMID:11572776.
- Orlando D, Lin C, Bernard A, Wang J, Socolar J, Iversen, E, Hartemink A, Haase SB. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature. 2008;453(7197):944–7. doi:10.1038/nature06955 doi:10.1038/nature06955. PMID:18463633.
- Simmons Kovacs L, Mayhew M, Orlando D, Jin Y, Li Q, Huang C, Reed S, Mukherjee S, Haase SB. Cyclin-dependent kinases are regulators and effectors of oscillations driven by a transcription factor network. Mol Cell. 2012;45(5)669–79. doi:10.1016/j.molcel.2011.12.033. doi:10.1016/j.molcel.2011.12.033. PMID:22306294.
- Bristow S, Leman A, Simmons Kovacs L, Deckard A, Harer J, Haase SB. Checkpoints couple transcription network oscillator dynamics to cell-cycle progression. Genome Biol. 2014;15(9):446. doi:10.1186/s13059-014-0446-7. doi:10.1186/s13059-014-0446-7. PMID:25200947.
- Cho-Yi C, Motta F, Kelliher C, Deckard A, Haase SB. Reconciling Conflicting Models for Global Control of Cell-Cycle Transcription. Cell Cycle. 2017;16(20):1965–1978. doi:10.1080/15384101.2017.1367073. PMID:28934013.
- Rahi S, Pecani K, Ondracka A, Oikonomou C, Cross FR. Cell. 2016;165(2):475–87. doi:10.1016/j.cell.2016.02.060.
