ABSTRACT
An accumulated evidence supports that MicroRNAs (miRNAs) have shown a prominent role in pathological processes and different tumor onset. However, to date, the potential functional roles and molecular mechanisms by how microRNA-424-5p(miR-424-5p) affects cancer cell proliferation are greatly unclear, especially in epithelial ovarian cancer(EOC).In this study, we demonstrated that miR-424-5p was significantly down-regulated in EOC tissues and cell lines. The level of miR-424-5p was negatively correlated with tumor size, TNM stage, pathological grade, lymphatic metastasis of EOC. Restoring miR-424-5p expression in EOC cells dramatically suppressed cell proliferation and caused an accumulation of cells in G1 phase, and thus contributed to better prognosis of EOC patients. Mechanistically, miR-424-5p inhibits CCNE1 expression through targeting CCNE1 3′UTR, and subsequent arrest cell cycle in G1/G0 phase by inhibiting E2F1-pRb pathway. This study revealed functional and mechanistic links between miR-424-5p and CCNE1 in the progression of EOC and provide an important insight into that miR-424-5p may serve as a therapeutic target in EOC.
Introduction
Ovarian cancer is the fifth most common cancer and the most malignant gynecological tumor seriously threatening females' lives and health worldwild. It caused 22,280 new diagnosed cases and accounted for over 14,240 deaths of the year 2016 in USA [Citation1]. Ovarian cancer is a heterogeneous disease which can be subdivided into three main histologic subtypes: sex cord stromal tumors (including granulose, theca, and hilus cells), germ cell tumors (oocytes), and epithelial ovarian cancer (EOC). EOC accounting for 90% of ovarian malignancies is believed to arise from epithelial cells covering the outer surface of the ovary and to be stuck with 30% of 5-year survival rate [Citation2]. Moreover, despite an improving satisfactory cytoreductive surgery and platinum-based chemotherapy in ovarian cancer, survival rate of EOC has changed little over the past 30 year [Citation3], with relapse rate at the advanced stages of 75% high [Citation4]. Thus, finding specific or practical laboratory markers for early diagnosis of EOC pose a critical cancer research challenge in clinical practice.
MicroRNAs (miRNAs) are small non-coding RNA, approximately 22 nt nucleotides in length, and play a critical role as discrete gene regulatory switches by mRNA cleavage or translational inhibition or degradation [Citation5]. In recent years, an accumulated evidence supports that miRNAs have shown a prominent role in pathological processes and different tumor onset, including EOC, by altering expression of specific targets and fine-tuning of signaling pathways [Citation6–8], MiR-424-5p is presented in a cluster of miRNAs on human chromosomal location Xq26.3 [Citation9] and dentified in many tumor types. The expression levels of miR-424-5p were decreased in hepatocellular carcinoma [Citation10], esophageal squamous cell carcinoma [Citation11], and cervical cancer [Citation12], compared to the overexpression in pancreatic cancer [Citation13], which elucidated that miRNAs performance acting as oncogenes or tumor suppressors varies in different tissues. Although genome-wide integrative study of miRNAs expression profiling studies showed that miR-424-5p is downregulated in ovarian carcinoma [Citation14], the detailed regulation mechanism of miR-424-5p in EOC remains unkown.
In this study, we confirmed that miR-424-5p expression was downregulated in surgically excised EOC tissues as well as cell lines and associated with the clinicopathological parameters of EOC patients. Our findings provided evidence that miR-424-5p was a tumor suppressor gene in EOC. Furthermore, miR-424-5p regulated the proliferation, cell cycle of ovarian cancer by targeting CCNE1 through E2F1-pRB pathway in vitro. Taken together, it might be exploited for a therapeutic intervention by upregulating miR-424-5p in epithelial ovarian cancer.
Results
miR-424-5p was depressed in EOC and associated with clinicopathological features of EOC patients
To identify the role of miR-424-5p in EOC development, we used qRT-PCR to test miR-424-5p expression in 83 EOC tissues as well as 19 matched normal ovarian tissues. As shown in (a), compared with normal ovarian tissues, miR-424-5p was significantly decreased in cancer samples (p<0.01). Next, we analyzed miR-424-5p expression level in different histological subtypes of EOC including HGSC (high grade serous carcinoma), LGSC (low grade serous carcinoma) and other types (8 mucinous carcinoma, 2 endometrioid carcinoma, 1 clear cell carcinoma), founding that miR-424-5p expression level was significantly downregulated between HGSC and LGSC ((b), p<0.01). Interestingly, clinicopathological analyses showed that miR-424-5p downregulated expression was strongly associated with poor differentiated pathological grade (p = 0.0334), advanced FIGO stage (p = 0.0036), residual tumor size (p = 0.0174), and lymphatic metastasis (p = 0.0398) as shown in . However, we did not observe any obvious links between miR-424-5p levels and the patients age, ascites and serum CA125 level (all p>0.05). qRT-PCR revealed that miR-424-5p expression were all significantly lower in ovarian cancer cell lines (HO8910, SKOV3, A2780) compared to an immortalized ovarian epithelial cell linenormal cell line (HOSEpiC) ((c), all p<0.01). Furthermore, miR-424-5p expression revealed that down-regulated miR-424-5p was remarkably associated with poor prognosis in 83 EOC patients using overall survival (OS) ((d), p = 0.012) and recurrence-free survival (RFS) Kaplan-Meier survival analysis ((e), p = 0.020) .These results demonstrated that miR-424-5p expression was depressed in both human EOC tissues and ovarian cancer cell lines, and its downregulation was correlated with pathogenic condition and prognosis of EOC, which indicates that miR-424-5p may play a vital role in EOC progression and development.
Figure 1. The qRT-PCR analysis of miR-424-5p expression in EOC tissues and cell lines. (A) The expression of miR-424-5p was significantly downregulated in 83 EOC tissues, compared to normal ovarian tissues (##p<0.01). (B) The downregulation of miR-424-5p significantly differ between HGSC and LGSC in different histological subtypes of EOC. (##p<0.01). (C) The expression levels of miR-424-5p in ovarian cancer cell lines were all significantly lower than in the normal cell line. Data were shown by mean±SEM from three independent experiments, where appropriate, ##p<0.01. (D) Overall or (E)relapse-free Kaplan-Meier survival analysis for miR-424-5p expression revealed that down-regulated miR-424-5p is associated with poor prognosis in 83 EOC patients (p = 0.012, p = 0.020).
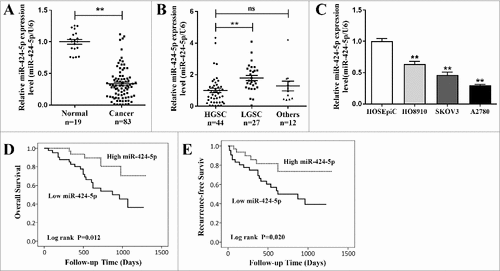
Table 1. Correlation between miR-424-5p expression and clinicopathological features in 83 EOCpatient.
Overexpression of miR-424-5p levels can induce EOC cells growth inhibition and arrest cell cycle progression in vitro
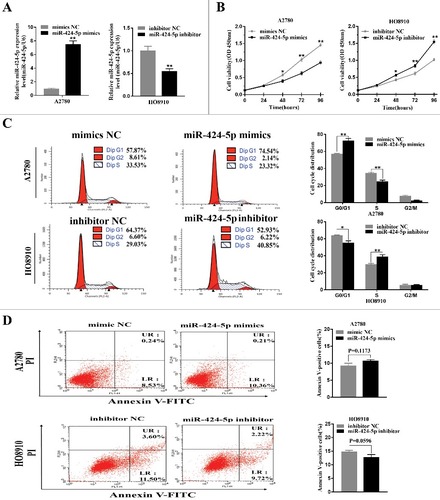
To better understand the biological functions of miR-424-5p, we used miR-424-5p mimics, inhibitor and NCs for transient transfection in the highest and lowest miR-424-5p expression cell lines, HO8910 and A2780 for further study. The transfection efficiencies for miR-424-5p overexpression or downregulation were confirmed by qRT-PCR after 24h ((a), p<0.01). CCK-8 assay was used to observe the impact of miR-424-5p on the cell proliferation and revealed that miR-424-5p suppressed EOC cell growth in a time-dependant manner. After 48h, miR-424-5p overexpression significantly inhibited cell growth in A2780 and downregulation of miR-424-5p remarkably promoted cell growth in HO8910 ((b), all p<0.05). Given that miR-424-5p suppresses the growth of EOC cells, we asked whether this inhibition may be caused by a block at a certain checkpoint in the cell cycle. As (c) shown above, cell cycle distribution analysis by flow cytometry in A2780 cell lines after 48h transfection demonstrated that ectopic miR-424-5p mimics cells had a signifcant increase in the percentage of cells G1/G0 peak (p<0.01) and a remarkable decrease in the G2/M phase (p<0.01),compared with its NC group. While a significant decrease in the G2/M phase (p<0.05) and remarkable increase in G1/G0 phase (p<0.01) can be seen in miR-424-5p inhibitor transfected HO8910 cell line, compared to its inhibitor NC group. These results suggested that miR-424-5p could trigger cell proliferation inhibition by arresting the tumor cells at the G1/G0 phase. We also assessed the effect of miR-424-5p on apoptosis in EOC cells. However, our data indicated that there was no difference in apoptosis between the miR-424-5p mimics, inhibitor and its NC groups in A2780 and HO8910 cell lines ((d), p = 0.1173, 0.0596, respectively).
Figure 2. Effect of miR-424-5p on cellular proliferation and cycle distribution. (A) Levels of miR-424-5p in A2780 and HO8910 cells were analyzed 24 h after mimics or inhibitor transfection. (B) Overexpression of miR-424-5p suppresses EOC cell growth. Cell viability was measured by CCK-8 assay on 24, 48, 72, 96 hours after miR-424-5p mimics, inhibitor and its controls transfected in A2780 and HO8910 cells, respectively. (C) Upregulation of miR-424-5p promotes G1/S transition in A2780, and downregulation of miR-424-5p arrests G1/S transition in HO8910, compared with the corresponding controls. (D) No significant difference between A2780 and HO8910 each treated with miR-424-5p mimics, inhibitor and its control groups in apoptosis. The data were shown by mean±SEM from three independent experiments, where appropriate, #p<0.05; ##p<0.01.
miR-424-5p directly targets CCNE1 3'-UTRs
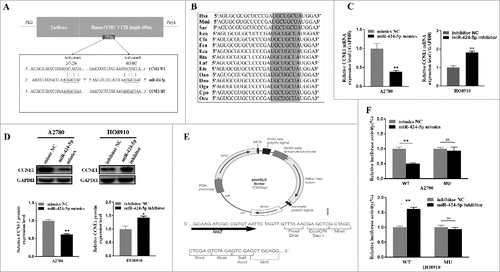
To explore the mechanisms of miR-424-5p suppressing tumor cell proliferation, cell cycle distribution, we first performed three well-known bioinformatics prediction algorithms by TargetSCAN, microRNA and miRDB. There were three candidate target genes for miR-424-5p (SLC9A6,ANO3,CCNE1)and CCNE1 was confirmed potential target gene of miR-424-5p by qRt-PCR in EOC cell line (Supplementary Figure 1). CCNE1 3′-UTR contains two putative binding sites (UGCUGCU) for the miR-424-5p seed sequence (AGCAGCA) which were predicted to be located at nt 247–254 (named target site A) and nt 485–492 (named target site B) respectively ((a)). More importantly, both of them are highly conserved in different species ((b)). Furthermore, mRNA and protein expression level of CCNE1 was significantly upregulated in A2780 cells transfected with miR-424-5p mimic, whereas downregulated in HO8910 cells transfected with miR-424-5p inhibitor ((c,d), #p<0.05, #p<0.05 ). Next, we generated a series of dual luciferase reporter plasmids to investigate the interaction between miR-424-5p and its predicted target sites within CCNE1 3′UTR ((e)). We cloned the human CCNE1 3′-UTR into a luciferase reporter plasmid and co-transfected it with miR-424-5p mimics and inhibitor into A2780 and HO8910 cells, respectively. Dual-luciferase reporter assays showed that miR-424-5p dramatically impeded the activity of firefly luciferase with wild construct (CCNE1 3′-UTR WT) ((f), all p<0.01), while this inhibitive effect was abolished when the reporter vectors contained a mutant form (CCNE1 3′-UTR MU). These results showed that miR-424-5p may restrain CCNE1 expression by directly binding to its 3′-UTR.
Figure 3. CCNE1 is a direct target gene of miR-424-5p in vitro. (A) Schematic representation of the CCNE1 3′-UTR with two putative sites (Seed location A and B) targeted by miR-424-5p. The sequence of miR-424-5p target sites in CCNE1 3′-UTR is shown in underlined. (B) Sequence alignment of predicted miR-424-5p target sites located within CCNE1 3′-UTR showing high conservation among different species. (C) Relative mRNA and protein expression of CCNE1 were confirmed by qRT-PCR and western blot when transfected miR-424-5p mimics and inhibitor each into A2780 and HO8910,compared with its controls. (E) Structure of pmiR-RB-Report^TM dualluciferase reporter vector. (F) Relative Lucuciferase activity was assayed and calculated by the ratio of firefly/Renilla luciferase activity following transfection with miR-424-5p mimic compared with transfection with NC in A2780, or transfection with miR-424-5p inhibitor compared with transfection with NC inhibitor in HO8910. The data were shown by mean±SEM from three independent experiments, where appropriate, #p<0.05; ##p<0.01.
CCNE1 is involved in the antitumor effects of miR-424-5p
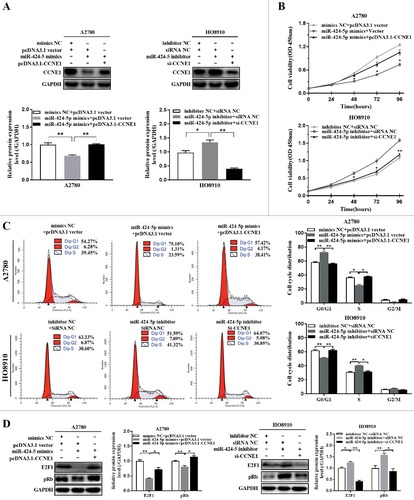
In order to test whether exogenously expressed CCNE1 (without 3’-UTR) could influence the suppressive effect of miR-424-5p on cell proliferation and cell cycle distribution or not, firstly, relative CCNE1 expression in both pcDNA3.1-CCNE1 co-transfected with miR-424-5p mimics A2780 cells and CCNE1 silencing with miR-424-5p inhibitor transfected HO8910 cells were confirmed by western blot, the result revealed that re-expression of CCNE1 in A2780 cells transfected with miR-424-5p mimics reversed the decrease of CCNE1 expression suppressed by miR-424-5p, compared with vector controls. Knockdown of CCNE1 in HO8910 cells transfected with miR-424-5p inhibitor promoted the decrease of CCNE1 expression induced by miR-424-5p, compared with siRNA controls ((a), #P<0.05, ##P<0.01). Furthermore, overexpression of CCNE1 dramatically restored the inhibitory effects of miR-424-5p in A2780 cell proliferation, whereas knocking down CCNE1 remarkably enhanced the inhibitory effects of miR-424-5p in HO8910 cell proliferation ((b), #P<0.05, ##P<0.01). Flow cytometry analysis indicated that ectopic expression of CCNE1 significantly counteracted cell cycle G1 phase arrest in miR-424-5p overexpression A2780 cells, and Knockdown of CCNE1 remarkably increased cell cycle G1 phase arrest in miR-424-5p downregulated HO8910 cells ((c), #P<0.05, ##P<0.01). Together, all the results above provided a further evidence in support of CCNE1 may act as an important functional target for miR-424-5p in EOC cells. Our result also found that introduction of miR-424-5p mimics significantly induced an inhibition of pRb and supressed E2F1 expression, while knockdown of miR-424-5p had the opposite effect on cell proliferation ((d)). These results indicated that miR-424-5p also downregulates CCNE1-mediated Rb phosphorylation and involves in inhibiting the activation of the E2F1–pRb signaling pathway.
Figure 4. Re-expression or knockdown CCNE1 influnced the suppressive effect of miR-424-5p on cell viability and cell cycle distribution. (A) Western blot analysis of CCNE1 protein level was examined in miR-424-5p overexpressing or downregulated in A2780 and HO8910 cells transfected with pcDNA3.1-CCNE1 and si-CCNE1, respectively. (B) The cell viability was assessed using CCK-8 assay after overexpression and silencing CCNE1 on miR-424-5p mimics-transfected A2780 and miR-424-5p inhibitor-transfected HO8910 cells for 0 h, 24 h, 48 h, 72 h and 96 h. (C) Cell cycle distributions of cells transfected with pCMV-CCNE1 or si-CCNE1 and their control groups were assessed by flow cytometry. The data were shown by mean±SEM from three independent experiments, where appropriate, #p<0.05; ##p<0.01, compared with its vector group or siRNA NC group. (D) Representative images for western blot were shown to analyze the relative protein expression of E2F1 and pRb after pCMV-CCNE1 or si-CCNE1 transfection each on miR-424-5p overexpressed A2780 and miR-424-5p downregulated HO8910 cells for 48h. GAPDH was used for normalization. The data were shown by mean±SEM from three independent experiments, where appropriate, #p<0.05; ##p<0.01, compared with its vector group or siRNA NC group.
miR-424-5p was inversely correlated with CCNE1 expression in EOC tissues
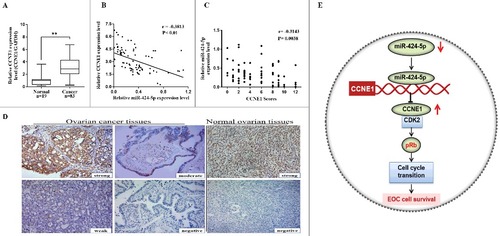
To further explore the relationship between miR-424-5p and CCNE1 in EOC patient tissues, immunohistochemistry assay and qRT-PCR were used to analyze the expression of CCNE1 in all clinical specimens. The result showed that CCNE1 mRNA expression level and IHC scores were both significantly higher in 83 EOC tissues than that in 19 corresponding normal tissues ((a), ##p<0.01). Moreover, pearson correlation analysis revealed that there was a statistically significant inverse correlation between miR-424-5p and CCNE1 in mRNA level ((b), r = −0.3813, p<0.01). As different intensity score immunohistochemical analysis of CCNE1 shown in (d), IHC scores revealed that CCNE1 was negatively associated with miR-424-5p expression ((c), r = −0.3143, p = 0.0038). These results revealed that CCNE1 expression in EOC tissues was inversely correlated with miR-424-5p levels ((e)).
Figure 5. Inverse correlation between miR-424-5p and CCNE1 expression in EOC tissues. (A) CCNE1 mRNA expression level was remarkably higher in 83 EOC tissues than in 19 corresponding normal tissues (##p<0.01). (B) miR-424-5p expression level was inversely correlated with CCNE1 mRNA expression in EOC tissues (n = 83, r = −0.3813, ##p<0.01) by Spearman's correlation analysis. (C) The staining score of CCNE1 inversely correlated with miR-424-5p levels in 83 EOC tissues. (Spearman's correlation analysis, r = −0.3143, p = 0.0038). (D) Representative immunohistochemical analyses of CCNE1 in normal ovarain tissues and EOC tissue with different intensity score (strong, moderate, week, negative) (X200). (E) Schematic depicting the regulation of cell survival by miR-424-5p in epithelial ovarian cancer.
Discussion
miRNAs are proved to be associated with gene regulation and varieties of tumorigenic processes, including cell proliferation [Citation15], apoptosis [Citation16], migration [Citation17], and invasion [Citation18] by multiple studies. The investigation of miRNA-mRNA interactions has drawn great attention from bench to bedside to understand their roles in tumor biology [Citation19]. The expression patterns of miRNAs in tumors have been proposed as biomarkers for diagnosis or prognostic determination and therapeutic intervention [Citation20]. Previous study found that ectopic expression of miR-424-5p is upregulated significantly in gastric cancer [Citation21], oral squamous cell carcinoma [Citation22], and colorectal cancer [Citation23]. However, the expression of miRNA-424-5p was found to be downregulated in hepatocellular carcinoma [Citation24], breast cancer tumor [Citation25] and cervical cancer [Citation26]. These paradoxical results may reflect the complexity of function of miR-424-5p as oncomiRs or tumor suppressors in different cancer types. Mechanistic studies have reported that overexpression of miR-424-5p can inhibite cell proliferation and promote cervical cancer cell apoptosis by targeting KDM5B-Notch pathway [Citation12]. In hepatocellular carcinoma, miR424-5p was down-regulated and suppressed cell migration and invasion by targeting c-Myb [Citation27].
In the revision time of this study, Wu [Citation28] has reported that miR-424-5p was downregulated in 30 tumors compared with adjacent non-tumor tissues. miR-424-5p inhibited malignant behaviors, invasive phenotype and EMT by downregulating doublecortin-like kinase 1 (DCLK1) in a special type of EOC, ovarian clear cell carcinom, which was consistent with our data, and supported our proposal that miR-424-5p may act as a suppressor of ovarian cancer. Our data have demonstrated that miR-424-5p was frequently down-regulated in EOC cell lines and tissues and low expression of miR-424-5p was associated with the aggressive clinical characteristics and poor prognosis in EOC patients.
There was another study on Senile Hemangiom revealing that CCNE1 was also one of predicated targets for miR-424-5p [Citation29]. Yet for all that, the effect of miR424-5p remains to be illuminated to supplement the network of its interactions in ovarian cancer. In our present study, we found that miR-424-5p was significantly repressed in EOC tissues and ovarian cancer cell lines. Overexpression of miR-424-5p decreased the proliferation, tumorigenicity and cell cycle progression in ovarian cancer cells. Moreover, we demonstrated that restoration of miR-424-5p inhibited ovarian cancer cells proliferation by inducing G1-S arrest but did not influence apoptosis of EOC cells probably because the strong vitality of cancer cells, which suggested that miR-424-5p regulates cell proliferation mainly through modulate cell cycle progression in EOC cells.
To investigate the downstream targets of miR-424-5p that may play a role in mediating its cell function, we searched for putative targets using some bioinformatics analyses available online. Through luciferase assays, we predicated miR-424-5p was directly targeting the CCNE1 3’-UTR in ovarian cancer cells. Previous studies have showed that CCNE1, a nuclear protein of cell cycle regulator family member, acts as an oncogene in cell cycle G1 progression and entry into S phase [Citation30]. It's reported that overexpression of cyclin E1 are related to various malignant tumors [Citation31–33], including ovarian cancers [Citation34]. Recently, accumulating evidence has indicated that expression of the CCNE1 can be regulated by miR-144-5p [Citation35], miR-16 [Citation36], and MicroRNA-30c-2-3p [Citation37] in other cancer cells. In our study, we confirmed that the protein expression of CCNE1 was post-transcriptionally down-regulated by overexpression of miR-424-5p using western blot analysis and involved in the antitumor effects of miR-424-5p through cell proliferation and cycle distribution. Further, we analyzed that miR-424-5p expression levels were significantly lower in CCNE1 high scores groups than those in low scores ones. The expression of miR-424-5p exhibited an inverse relationship with the expression of CCNE1 in EOC tissues. Additionally, we showed that overexpression of miR-424-5p led to the suppression of phosphorylated pRb and E2F1 through downregulation of cyclin E1. These findings support the prediction that CCNE1 is a new downstream target of miR-424-5p and can suppress activity of molecues in pRb-E2F1 pathway.
In summary, our findings indicate that miR-424-5p functions as a tumor suppressor and impedes proliferative and metastatic effects through blocking the G1/S phase transition in ovarian cells. Mechanistically, we confirm that miR-424-5p evoked G0/G1 cell cycle arrest by targeting CCNE1 mediated E2F1-pRb signaling pathway. Taken together, miR-424-5p may serve as a novel potential therapeutic target for EOC treatment.
Materials and methods
Clinical samples and characteristics
83 surgical resection of primary tumors and 19 matched normal tissues were immediately preserved by snap-frozen in liquid nitrogen and stored at −80°C from Department of Gynecology, Qilu Hospital of Shandong university (China) during 2015–2017. All patients were staged according to the 2009 international federation of gynecology and obstetrics (FIGO) staging guideline and histologically confirmed by two pathologist. The study was approved by Ethics Committee of Shandong University and all patients have signed informed consents. Before diagnosis, none of patients have received surgery or chemotherapy previously. The clinicopathologic characteristics parameters were collected in .
Cell lines and cell culture conditions
EOC cell lines (SKOV-3,HO8910,A2780) were obtained from the Cancer Center Laboratory of Shandong University (Jinan, China) and Normal Human Ovarian Surface Epithelial cells (HOSEpiC) cell line was purchased from Jennio Biotech company (Guangzhou, China). Cell lines except SKOV-3 which was supplemented with McCoy's 5A medium (Gibco, Sydney, Australia) were all cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Sydney, Australia) with 10% fetal bovine serum (Gibco, Sydney, Australia) and 1% penicillin-streptomycin (Invitrogen, USA) at 37℃ in a humidified chamber with 5% CO2.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cultured cell lines and fresh tissue samples with TRIzol reagent (Life Technologies, Carlsbad, CA, USA) according to manufacturer's instructions. The concentration and quality of total RNA was determined using NanoDrop 2000 Spectrophotomete (Thermo Scientific Inc, Waltham, MA, USA). 3–5μg of RNA with miR-424-5p, U6 or random hexamer RT primers were used to synthesize cDNA according to M-MLV reverse transcriptase instruction (Invitrogen, Shanghai, China). Products were amplified by PCR under the following conditions: 95°C for 5min, followed by 40 cycles at 95°C for 30s, 60°C for 30s and 72°C for 30s using the Quant one step SYBR Green qRT-PCR Kit (Tiangen, Beijing, China) on an ABI 7500 System. miRNA (hsa-miR-424-5p) and normalized control (sn-RNA U6) RT primers (not provided) were obtained from Genepharma (Shanghai, China). CCNE1, SLC9A6, ANO3 and GAPDH PCR primer-probe sets were designed referring the Primer Bank primers and synthesized by DINGGUO CHANGSHENG company (Beijing, China).All PCR primers sequence were listed in Supplementary Table 1. Relative expression levels of miRNA or mRNA were analyzed using the StepOnePlus™ software (Applied Biosystems, Shanghai, China) and quantified by the 2−ΔΔCt method.
Transient transfection
Cells were seeded into six-well plates at a density of 2 × 105 cells/well with 2 mL of medium without antibiotics and transfection was typically carried out on cells until they were 70–90% confluent. To achieve the transient overexpression or knockdown of miR-424-5p, the miR-424-5p mimic, inhibitor as well as matched controls (mimics nc, inhibitor nc) were purchased from Genepharma company (Shanghai, China) with their sequences listed in Supplementary Table 2. CCNE1 siRNA and its non-targeting control (siRNA NC) were purchased from Ambion (Invitrogen, CA, USA). pcDNA3.1-CCNE1 plasmid was constructed by GenePharma. Transfection was performed according to the Lipofectamine™ 2000 manufacturer's instructions (Invitrogen, Carlsbad, CA). Total RNA and protein were used for qRT-PCR or western blot analysis, respectively, after transfection 24–48h.
Western blot analysis
Total protein was extracted using RIPA lysis buffer (Beyotime Biotech, Haimen, China) with 1% phenylmethylsulfonyl fluoride (PMSF) and 1% NaF for 30 min. Cell lysates with equal protein (30-50 µg) were separated by 10%SDS–polyacrylamide gels and transferred to nitrocellulose membranes (Millipore, Darmstadt, Germany). After blocking with 5% milk in Tris-buffered saline plus 0.02% Tween-20 (TBST) for 1h at room temperature, membranes were then incubated overnight at 4˚C with primary rabbit or mouse monoclonal antibodies against CCNE1, E2F1, phosphor-Rb (pRb) and GAPDH (Supplementary Table 3). Species-specific horseradish peroxidase–conjugated (HRP) secondary antibody anti-rabbit and anti-mouse (Millipore, Darmstadt, Germany) were used for the detection of primary antibodies. Protein levels were detected by the ECL blotting detection (Beyotime, Beijing, China), visualized on ImageQuant LAS 4000 (GE Healthcare Life Sciences, Logan, UT, USA) and quantified with normalized GAPDH by ImageJ software (NIH, Bethesda, USA).
CCK8 assay
EOC cells were seeded at 2 × 103 per well in 96-well plates and cultured in mediums containing 10% FBS at 37°C and 5% CO2. To evaluate the cell proliferation Cell at monitored time point (24 h, 48 h, 72 h or 96h respectively), counting Kit-8 (CCK8) (Tongren, Shanghai, China) (10 µL) was added to each plate at 4 hours before assessing the optical density (OD)values at 450 nm on a scanning multi-well spectrophotometer (Infinite 2000, Tecan, Männedorf, Switzerland).
Luciferase reporter assay
Bioinformatic target genes of miR-424-5p were predicted using 3 online miRNA target databases interseciton: TargetSCAN, Microrna and miRDB. CCNE1, one of candidate target genes, was containing two putative binding seed sites (247–254, 485–492) of miR-424-5p sequences in the 3′-UTR complementary and was amplified as well as mutant sequences by PCR into a pGL3-control vector (Promega, Madison, WI, USA) with the Xba I site). EOC cells were seeded at 1.5 × 105 cells per well in a 24-well plate. Wild-type CCNE1-3′-UTR (100 ng) and mutant CCNE1-3′-UTR (100 ng) were co-transfected with miR-424-5p mimics or the negative control (50 nM) using Lipofectamine 2000 (Invitrogen) for luciferase reporter assays. Twenty-four hours after transfection, cells were collected for frefly and Renilla luciferase activities detection using the Dual-Luciferase Reporter Assay System.
Immunohistochemistry staining and evaluation
4-µm sections were cut and deparaffinized, rehydrated, and autoclaved at 121°C by microwave heating for 15 minutes for antigen retrieval. Endogenous peroxidase was blocked with 3% hydrogen peroxide for 10 minutes. Antibody anti-CCNE1 (1:500 dilution in PBS, Abcam, Cambridge, UK) and a Polink-2 Plus Polymer HRP Detection System (ZSGB-BIO, Beijing, China) were used for primary antibodies incubation overnight at 4°C. The nucleus was counterstained using Meyer's hematoxylin ( Solarbio, Beijing, China). Sections were stained by diaminobenzidine (DAB, ZSGB-BIO, Beijing, China). Semiquantitative analysis of staining results was assessed by two blinded independent pathologists. The percentage of positively stained positive tumor cells was estimated and expressed as a proportional score (PS): <10% socores 0; 10–25% socores 1; 26–50% socores 2; 51–75% socores 3; >75% socores 4. An intensity score (IS) was also used for scoring the staining intensity and presented as negative = 0, weak = 1, moderate = 2, strong = 3. The total score (TS) = PS × IS (range 0–12). Finally, the CCNE1 expression cases were classified into two different groups: low (score 0–6) and high (score 8–12).The images were captured by the Olympus IX51 inverted microscope (Olympus, 8 Tokyo, Japan).
Flow cytometric Cell cycle and Apoptosis analysis
As for cell cycle analysis, transfected cells after centrifugation were washed 3 times with precooling phosphate-buffered saline (PBS), and then fixed in 70% cold ethanol overnight. According to the Cell Cycle Detection Kit (Leagene Biotechnology, Beijing, China) manufacturer's instruction, each sample was washed and resuspended using 500 μl propidium iodide (PI) working solution for 30 minutes in the dark before detection by fluorescence-activated cell sorting (FACS) Calibur flow cytometry (BD Biosciences, San Jose, CA, USA). The percentage of cells was further processed with the ModFit LT software (Tree Star, USA). As for apoptosis analysis, Annexin V–FITC/PI Apoptosis Detection Kit purchased from BIBOX company (Nanjing, China) were used to stain the apoptosis cells at room temperature for 10 minutes before analysis with a BD FACS Calibur flow cytometry.
Statistical analysis
All statistical analysis were performed by GraphPad Prism version 7.03 (GraphPad Software Inc, San Diego, CA, USA). The differences of two or three groups were used student's t-test or One-way ANOVA. Survival curves were plotted using the Kaplan–Meier method and compared with the log–rank test. Correlation analysis was analyzed by Pearson's two-tailed correlation coefficient analysis. The correlations between miR-424-5p expression and clinical-pathological parameters was determined using chis quare test. Data are shown as the mean±SEM from three separate experiments. Statistical tests were two-sided and considered statistically significant at p < 0.05.
Compliance with ethical standards
Our studies involving human participants has got the approval by Ethics Committee of Shandong University and were performed in accordance with the ethical standards laid down in the 1975 Declaration of Helsinki.
Disclosure of potential conflicts of interest
The authors declare that they have no conflicts of interest.
2017CC7789R_s01__1_.doc
Download MS Word (570 KB)Acknowledgments
We thank for Professor Jinbo Feng and Zhenping Liu from Gene Research Cente for technical assistance and Professor Beihua Kong's supplyment of cell lines. We appreciate Dr. Ming Yang's help for collecting tissues (Qilu Hospital, Shandong University).
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332. PMID:26742998.
- Bast RC Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9(6):415–428. doi:10.1038/nrc2644. PMID:19461667.
- Vaughan S, Coward JI, Bast RC Jr, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11(10):719–725. doi:10.1038/nrc3144. PMID:21941283.
- Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177(3):1053–1064. doi:10.2353/ajpath.2010.100105. PMID:20651229.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. 14744438 doi:10.1016/S0092-8674(04)00045-5. PMID:14744438.
- Ge T, Yin M, Yang M, et al. MicroRNA-302b suppresses human epithelial ovarian cancer cell growth by targeting RUNX1. Cell Physiol Biochem. 2014;34(6):2209–2220. doi:10.1159/000369664. PMID:25562167.
- Ying X, Wei K, Lin Z, et al. MicroRNA-125b suppresses ovarian cancer progression via suppression of the Epithelial-Mesenchymal transition pathway by targeting the SET protein. Cell Physiol Biochem. 2016;39(2):501–510. doi:10.1159/000445642. PMID:27383536.
- Chen X, Dong C, Law PT, et al. MicroRNA-145 targets TRIM2 and exerts tumor-suppressing functions in epithelial ovarian cancer. Gynecol Oncol. 2015;139(3):513–519. doi:10.1016/j.ygyno.2015.10.008. PMID:26472353.
- Wentz-Hunter KK, Potashkin JA. The role of miRNAs as key regulators in the neoplastic microenvironment. Mol Biol Int. 2011;2011:839872. doi:10.4061/2011/839872. PMID:22091413.
- Zhang Y, Li T, Guo P, et al. MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Sci Rep. 2014;4:6248. doi:10.1038/srep06248. PMID:25175916.
- Wang F, Wang J, Yang X, et al. MiR-424-5p participates in esophageal squamous cell carcinoma invasion and metastasis via SMAD7 pathway mediated EMT. Diagn Pathol. 2016;11(1):88. doi:10.1186/s13000-016-0536-9. PMID:27628042.
- Zhou Y, An Q, Guo RX, et al. miR424-5p functions as an anti-oncogene in cervical cancer cell growth by targeting KDM5B via the Notch signaling pathway. Life Sci. 2017;171:9–15. doi:10.1016/j.lfs.2017.01.006. PMID:28082020.
- Wu K, Hu G, He X, et al. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol Oncol Res. 2013;19(4):739–748. doi:10.1007/s12253-013-9637-x. PMID:23653113.
- Dong R, Liu X, Zhang Q, et al. miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget. 2014;5(21):10816–10829. doi:10.18632/oncotarget.2522. PMID:25333261.
- Chang RM, Xiao S, Lei X, et al. miRNA-487a Promotes proliferation and metastasis in hepatocellular carcinoma. Clin Cancer Res. 2016. doi:10.1158/1078-0432.CCR-16-0851. PMID:27827315
- Joshi P, Jeon YJ, Lagana A, et al. MicroRNA-148a reduces tumorigenesis and increases TRAIL-induced apoptosis in NSCLC. Proc Natl Acad Sci USA. 2015;112(28):8650–8655. doi:10.1073/pnas.1500886112. PMID:26124099.
- Sandbothe M, Buurman R, Reich N, et al. The microRNA-449 family inhibits TGF-beta-mediated liver cancer cell migration by targeting SOX4. J Hepatol. 2017;66(5):1012–1021. doi:10.1016/j.jhep.2017.01.004. PMID:28088579.
- Samaeekia R, Adorno-Cruz V, Bockhorn J, et al. miR-206 Inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 Pathway. Clin Cancer Res. 2017;23(4):1091–1103. doi:10.1158/1078-0432.CCR-16-0943. PMID:27435395.
- Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17(12):719–732. doi:10.1038/nrg.2016.134. PMID:27795564.
- Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34–49. doi:10.1016/j.pharmthera.2016.11.012. PMID:27916656.
- Wei S, Li Q, Li Z, et al. miR-424-5p promotes proliferation of gastric cancer by targeting Smad3 through TGF-beta signaling pathway. Oncotarget. 2016;7(46):75185–75196. doi:10.18632/oncotarget.12092. PMID:27655675
- Peng HY, Jiang SS, Hsiao JR, et al. IL-8 induces miR-424-5p expression and modulates SOCS2/STAT5 signaling pathway in oral squamous cell carcinoma. Mol Oncol. 2016;10(6):895–909. doi:10.1016/j.molonc.2016.03.001. PMID:27038552.
- Torres S, Garcia-Palmero I, Bartolome RA, et al. Combined miRNA profiling and proteomics demonstrates that different miRNAs target a common set of proteins to promote colorectal cancer metastasis. J Pathol. 2017;242(1):39–51. doi:10.1002/path.4874. PMID:28054337.
- Yang H, Zheng W, Shuai X, et al. MicroRNA-424 inhibits Akt3/E2F3 axis and tumor growth in hepatocellular carcinoma. Oncotarget. 2015;6(29):27736–27750. doi:10.18632/oncotarget.4811. PMID:26315541.
- Rodriguez-Barrueco R, Nekritz EA, Bertucci F, et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31(6):553–566. doi:10.1101/gad.292318.116. PMID:28404630.
- Xu J, Li Y, Wang F, et al. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32(8):976–987. doi:10.1038/onc.2012.121. PMID:22469983.
- Yao H, Liu X, Chen S, et al. Decreased expression of serum miR-424 correlates with poor prognosis of patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8(11):14830–14835. 26823812 PMID:26823812
- Wu X, Ruan Y, Jiang H, et al. MicroRNA-424 inhibits cell migration, invasion, and epithelial mesenchymal transition by downregulating doublecortin-like kinase 1 in ovarian clear cell carcinoma. Int J Biochem Cell Biol. 2017;85:66–74. doi:10.1016/j.biocel.2017.01.020. PMID:28161486.
- Nakashima T, Jinnin M, Etoh T, et al. Down-regulation of mir-424 contributes to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma: its implications to therapy. PLoS One. 2010;5(12):e14334. doi:10.1371/journal.pone.0014334. PMID:21179471.
- Dulic V, Lees E, Reed SI. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257(5078):1958–1961. 1329201 doi:10.1126/science.1329201. PMID:1329201.
- Nault JC, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47(10):1187–1193. doi:10.1038/ng.3389. PMID:26301494.
- Herrera-Abreu MT, Palafox M, Asghar U, et al. Early adaptation and acquired resistance to CDK4/6 Inhibition in estrogen receptor-positive breast cancer. Cancer Res. 2016;76(8):2301–2313. doi:10.1158/0008-5472.CAN-15-0728. PMID:27020857.
- Fu YP, Kohaar I, Moore LE, et al. The 19q12 bladder cancer GWAS signal: association with cyclin E function and aggressive disease. Cancer Res. 2014;74(20):5808–5818. doi:10.1158/0008-5472.CAN-14-1531. PMID:25320178.
- Karst AM, Jones PM, Vena N, et al. Cyclin E1 deregulation occurs early in secretory cell transformation to promote formation of fallopian tube-derived high-grade serous ovarian cancers. Cancer Res. 2014;74(4):1141–1152. doi:10.1158/0008-5472.CAN-13-2247. PMID:24366882.
- Matsushita R, Seki N, Chiyomaru T, et al. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br J Cancer. 2015;113(2):282–289. doi:10.1038/bjc.2015.195. PMID:26057453.
- Guo X, Connick MC, Vanderhoof J, et al. MicroRNA-16 modulates HuR regulation of cyclin E1 in breast cancer cells. Int J Mol Sci. 2015;16(4):7112–7132. doi:10.3390/ijms16047112. PMID:25830480.
- Shukla K, Sharma AK, Ward A, et al. MicroRNA-30c-2-3p negatively regulates NF-kappaB signaling and cell cycle progression through downregulation of TRADD and CCNE1 in breast cancer. Mol Oncol. 2015;9(6):1106–1119. doi:10.1016/j.molonc.2015.01.008. PMID:25732226.
