ABSTRACT
The incidence and mortality rate of colorectal cancer (CRC) have been significantly increasing. However, mechanisms involved in CRC progression are still unclear. LncRNA ZFAS1 has been verified as oncogenic molecular in a series of tumors, including CRC. However, the underlying mechanism of ZFAS1 in CRC carcinogenesis remains unclear. In the present study, our data showed that ZFAS1 expression was significantly upregulated in CRC tissues and cell lines. Correlation analysis showed that high ZFAS1 expression was significantly associated with Helicobacter pylori infection, lymph nodes metastasis, advanced TNM stage and poor overall survival of CRC patients. Loss-of-function experiments revealed that ZFAS1 inhibition could markedly suppress CRC cells proliferation and invasion both in vitro and in vivo. Bioinformatics analysis and luciferase reporter assay revealed that ZFAS1 directly interacted with miR-484. Rescue experiments showed that miR-484 inhibitor reversed the tumor suppressing roles of ZFAS1 knockdown on CRC cells. Therefore, our study suggested that ZFAS1 could act as an oncogene in CRC tumorigenesis, and discovered the functional regulatory pathway of ZFAS1 sponging miR-484.
Introduction
Colorectal cancer (CRC) is one of the most common malignancies worldwide. Approximately 1.2 million patients worldwide have been diagnosed with CRC [Citation1,Citation2]. Although aggressive therapeutic strategies including surgery, radiotherapy, and chemotherapy have greatly improved, CRC morbidity and mortality remain high [Citation3]. Therefore, a better understanding of the molecular mechanisms involved in CRC will provide novel diagnostic biomarkers and effective therapeutics for the treatment of CRC patients.
Long non-coding RNAs (lncRNAs) are RNA molecules whose transcripts are more than 200 nt in length [Citation4]. They do not encode proteins, but play an important role in regulating gene expression through both epigenetic and posttranscriptional mechanisms [Citation5,Citation6]. Aberrant expression levels of lncRNAs have been correlated with various malignant biological processes, including carcinogenesis, cell proliferation, migration, invasion and apoptosis [Citation7,Citation8]. Increasing studies showed that lncRNAs play critical roles in tumor progression. For example, Lin et al showed that increased expression of the lncRNA ANRIL promoted lung cancer cell metastasis and correlates with poor prognosis [Citation9]. Li et al suggested that overexpression of lncRNA HOTTIP increased chemoresistance of osteosarcoma cell by activating the Wnt/β-catenin pathway [Citation10]. Wang et al indicated that lncRNA XIST exerted oncogenic functions in human glioma by targeting miR-137 [Citation11]. Thus, emerging evidences are proving the more and more significant role of lncRNA in tumor progression.
LncRNA zinc finger antisense 1 (ZFAS1) is an antisense transcript from 5’ end of Znfx1 gene, being firstly identified to play a functional role in patients with acute myocardial infarction [Citation12]. Moreover, the dysregulated ZFAS1 expression in cancer tissues has been discovered. For example, Gao et al found that lncRNA ZFAS1 could exhibit an oncogenic role in glioma progression by regulating EMT and Notch signaling pathway [Citation13]. Xia et al showed that lncRNA ZFAS1 interacted with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy [Citation14]. Recent studies showed that ZFAS1 play important roles in CRC. For example, Wang et al showed that upregulation of lncRNA ZFAS1 predicted poor prognosis and promoted invasion and metastasis in colorectal cancer [Citation15]. Thorenoor et al indicated that ZFAS1 interacted with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer [Citation16]. However, the underlying mechanism of ZFAS1 on CRC progression remains unclear.
In the present study, our data elucidated the regulatory function of ZFAS1 in the development of CRC via regulating miR-484, suggesting an effective therapeutic strategy for CRC treatment.
Materials and methods
Tissue specimens and ethics statement
The CRC tissues and adjacent non-tumor tissues were collected from 49 patients who underwent surgical resection at Huaihe Hospital of Henan University (China) between 2012 and 2013. All patients did not receive any local or systemic treatment before surgery. All the CRC tissues were confirmed by two experienced pathologists. All patients signed the informed consent and the experimental protocol was approved by the Ethics Committee of Huaihe Hospital of Henan University.
Cell lines and cell transfection
Human CRC cell lines (HCT116, SW480, SW620, HT-29 and LOVO) and the human colonic epithelial cells HCoEpiC were obtained from the American Type Culture Collection. All cells were cultured in RPMI-1640 medium (Gibco) supplemented with 10% FBS, 100 U/mL streptomycin and 100 U/mL penicillin (Gibco). The culture condition was at 37 °C in a humidified atmosphere containing 5% CO2.
For RNA interference assay, cells were seeded at 70–80% confluency before transfection and transfected with siRNAs using Lipofectamine 2000 (Invitrogen). All the siRNAs (20nM) were synthesized by Genepharma (Shanghai, China) and the sequences were as follows: si-ZFAS1-1, 5′-CUGGCUGAACCAGUUCCACAAGGUU-3′; si-ZFAS1-2, 5′-CCCTGTGCTTTCATGAAAGTGAAGA-3′; si-NC, 5′-CCAAAACCAGGCUUUGAUUGA-3’.
Total RNA extraction and quantitative real-time PCR
Tissues and cells samples were dissolved in TRIzol reagent (Invitrogen) and total RNAs was extracted according to the manufacturer's instruction. The cDNA was synthesized with the Reverse Transcription System (Promega). RT-PCR was performed using the SYBR Green Master Mixture (Roche) reagent in ABI 7500 Real-time PCR instrument. U6 was acted as an internal control. Relative expression levels were calculated and normalized using the 2−ΔΔC tmethod. All the primers were listed as follows: ZFAS1 forward, 5′-AAGCCACGTGCAGACATCTA-3′, reverse, 5′ CTACTTCCAACACCCGCATT-3′; miR-484, forward, 5′-CTCAACTGGTGTCTTCAGTTGAGGGTGGAGTCGGCC-3′, reverse 5′-ACACTCCAGGGCGCCGCGCTGGGAGGCGG-3′; U6, forward, 5′-CGCTAGCACATATCGGCTA-3′, reverse, 5′-TTCTGCGACGAATTTGTCAT-3′.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8, Dojindo) was employed to determine cell proliferation. Briefly, 1 × 104 cells were seeded into 96-well plates and 10 μl CCK-8 solution was added to each well. Then, the cells and plates were incubated at 37°C for 90 min. At the indicated time points (24, 48, 72 and 96 h), the absorbance at 450 nm was measured using a spectrophotometer.
Colony formation assay
For the colony formation assay, cells were seeded into six-well plates at density of 1 × 103 per well. After being cultured for 14 days, cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 15 min. then, cells were stained with crystal violet for 10 min and washed with PBS to remove the staining solution. Colonies were examined and counted under microscope. The assays were repeatedly performed in triplicate.
Transwell invasion assay
The cell invasion assay was performed using 24-well insert transwell chambers (8µm, Corning). Cells (2 × 104) suspended in 200 µL serum-free medium were added to the upper chamber, and culture medium containing 20% FBS was placed in the bottom chamber. The cells were then incubated for 48 h at 37°C; the cells on the upper surface were scraped and washed away, whereas the cells on the lower surface were fixed with 20% methanol and stained with 0.1% crystal violet. The number of invaded cells was counted in 5 randomly selected fields under a microscope.
Xenograft nude mouse model
Male BALB/c nude mice (6 weeks) were maintained in clean conditions and used for xenograft assay. cells (4 × 105/100ml) were transfected with sh-ZFAS1 or control (sh-NC), and then subcutaneously injected into back of nude mice. After that, tumor size was measured every week, and tumor weight was measured after sacrifice of mice. The xenograft mice assay was approved by the Committee on Animal Welfare of Huaihe Hospital of Henan University.
Luciferase reporter assay
For luciferase assay, the 3’-UTR of ZFAS1 cDNA was amplified using PCR, then, cDNA were cloned into the downstream of the firefly luciferase gene pGL3 (Invitrogen). HEK293T cells were seeded into 96-well plates at density of 1 × 104 cells/well. Afterwards, pGL3-ZFAS1-Wt (wild type) and pGL3-ZFAS1-Mut (mutant) was transfected into HEK293T cells with miR-484 mimics using Lipofectamine 2000 according to the manufacturer's instructions. After 24 h of transfection, luciferase activities were measured using the dual-luciferase reporter gene assay kit (Promega) according to the manufacturer's instructions.
RNA immunoprecipitation
Cells were lysed using a complete RNA lysis buffer containing protease inhibitor and RNase inhibitor using an EZ-Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) following the manufacturers’ instructions. 100uL of whole cell lysate was incubated with the RIP immunoprecipitation buffer containing magnetic beads coated with Ago2 antibody and was designated as the test group, while the control group consisted of normal mouse IgG. After being incubated for 2 h at 4°C, the immunoprecipitated RNA was isolated. The purified RNA was further used in the qRT-PCR analysis of ZFAS1 and miR-484.
Statistical analysis
All data were represented as mean ± standard deviation (SD) and statistical analyses were performed using SPSS 19.0. The differences were evaluated by the Student t-test and one way ANOVA. Pearson correlation analysis was used to estimate the relationship between the expression level of ZFAS1 and miR-484. Kaplan–Meier methods with the log-rank test were performed to calculate overall survival. p<0.05 was considered statistically significant.
Results
LncRNA ZFAS1 was upregulated in CRC tissues
To explore the clinical relevance of lncRNA ZFAS1 in human CRC. We detected ZFAS1 expression in CRC tissues using qRT-PCR. The results showed that ZFAS1 expression was significantly upregulated in CRC tissues compared to adjacent non-tumor tissues (A and B; p<0.05). According to the median value of ZFAS1 expression in CRC tissues, patients were divided into two groups: the high ZFAS1 expression group (n = 25) and low ZFAS1 expression group (n = 24). Results showed that high ZFAS1 expression was positively associated with Helicobacter pylori infection, lymph nodes metastasis and advanced TNM stage (; p<0.05). As shown in C, high ZFAS1 expression in CRC tissues is significantly associated with worse overall survival (p<0.05, log-rank test). In addition, ROC (Receiver Operating Characteristic) curve showed that comparing with CRC patients and controls, ZFAS1 had an AUC (Area under the Curve) value of 0.837 (95% CI: 0.713-0.921). Besides, sensitivity, specificity and p value were respectively 0.752, 0.788 and 0.006 (D). Taken together, those results indicated that ZFAS1 could act as an oncogene in CRC progression.
Figure 1. LncRNA ZFAS1 was upregulated in CRC tissues and associated with poor prognosis. (A,B) Relative expression of ZFAS1 measured by qRT-PCR in CRC tissues and adjacent non-tumor tissues. (C) Kaplan-Meier analysis and log-rank test were performed to assess the prognosis of CRC patients with high- and low- ZFAS1 expression. (D) ROC curve showed the AUC value of ZFAS1 (0.837, 95% CI: 0.713-0.921). *p<0.05
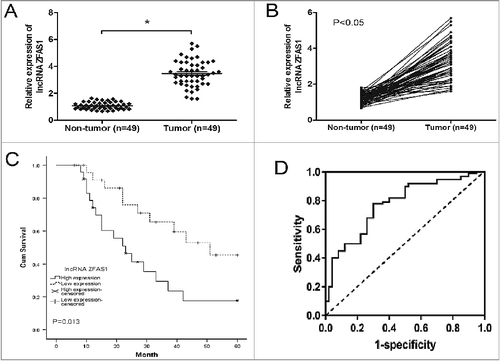
Table 1. Correlation between ZFAS1 expression and clinicopathological features of CRC.
ZFAS1 knockdown suppressed CRC cells proliferation and invasion in vitro
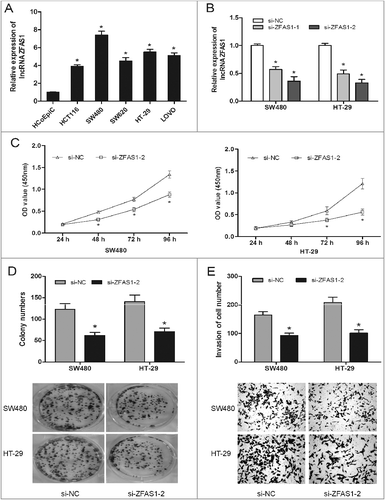
It had been testified that ZFAS1 was upregulated in CRC tissue, and the high ZFAS1 expression was closely correlated with advanced clinical features and poor overall survival. Thus, we performed loss-of-functional experiments to explore the role of ZFAS1 on CRC cells in vitro. QRT-PCR showed that ZFAS1 expression in CRC cells (HCT116, DLD1, SW480, SW620, HT-29 and LOVO) was significantly increased compared to HCoEpiC cells (A; p<0.05). The synthesized small interfering RNAs were respectively transfected into SW480 and HT-29 cells. The ZFAS1 expression was significantly decreased in si-ZFAS1 transfecting cells (B; p<0.05). CCK-8 assay showed that ZFAS1 knockdown inhibited cell viability in SW480 and HT-29 cells compared to si-NC group (C; p<0.05). Furthermore, colony formation assay showed that ZFAS1 inhibition effectively suppressed the clone formation ability of SW480 and HT-29 cells (D; p<0.05). Additionally, the invasion ability of SW480 and HT-29 cells transfected with si-ZFAS1 was inhibited compared with si-NC group (E; p0.05). Taken together, those results suggested that ZFAS1 suppression could inhibit CRC cell viability and invasion.
Figure 2. LncRNA ZFAS1 inhibition suppressed CRC cells proliferation and invasion. (A) Relative expression of ZFAS1 measured by qRT-PCR in CRC cells and human colonic epithelial cells HCoEpiC. (B) ZFAS1 expression was examined by qRT-PCR assay in cells transfected with si-ZFAS1 or si-NC. (C) CCK-8 assay showed that ZFAS1 depletion inhibited the proliferation of SW480 and HT-29 cells. (D) colony formation assay revealed that ZFAS1 inhibition suppressed the clone formation of SW480 and HT-29 cells. (E) Transwell invasion assay suggested that ZFAS1 suppression inhibited the invasion ability of SW480 and HT-29 cells. *p<0.05
ZFAS1 knockdown suppressed tumor growth in vivo
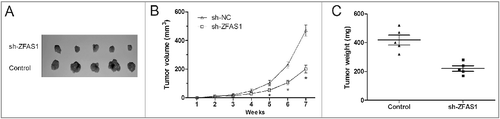
To confirm the biological function of ZFAS1 in CRC in vivo, SW480 cells stably transfected with sh-ZFAS1 or sh-NC were subcutaneously injected into the right flank of the nude mice. Tumor volume was measured every weeks for 7 weeks. As shown in A, ZFAS1 knockdown suppressed tumor growth in vivo compared with sh-NC group (p<0.05). Xenograft tumors were resected and weighed after 7 weeks of injection. Tumor weight in sh-ZFAS1 group was lower than that in sh-NC group (B and C; p<0.05). Thus, these data demonstrated that ZFAS1 knockdown could suppress tumor growth in vivo.
Figure 3. LncRNA ZFAS1 knockdown suppressed tumor growth in vivo. (A) SW480 cells transfected with sh-ZFAS1 were subcutaneously injected into back of nude mice. Tumor volume was measured every week. (B,C) The subcutaneous tumors were taken photos (B) and weighted (C) in sh-ZFAS1 group or sh-NC group. *p<0.05
miR-484 was directly regulated by ZFAS1
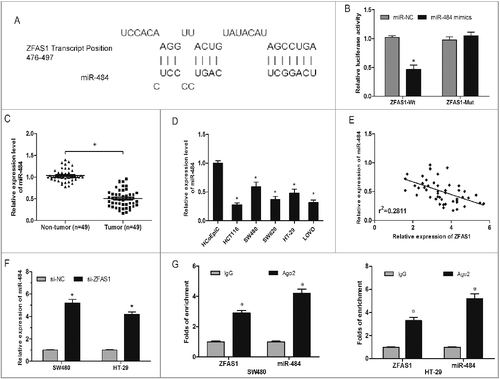
Recently, increasing studies demonstrated that lncRNA might act as a competing endogenous RNA (ceRNA) or a molecular sponge in regulating the biological functions of miRNAs [Citation17]. To investigate the underlying mechanism of ZFAS1 on CRC tumorigenesis, the bioinformatics tool DIANA was used to explore the targets of ZFAS1. Results showed that miR-484 shared complementary binding sites with ZFAS1 3’-UTR (A). Luciferase reporter assay revealed that miR-484 mimics could evidently suppress the luciferase activity of the ZFAS1-Wt, but not of ZFAS1-Mut (B; p<0.05). In addition, we explored the expression of miR-484 in CRC tissues and cell lines, results showed that miR-484 was significantly downregulated in CRC tissues and cell lines (C and D; p<0.05). Pearson's correlation analysis revealed an inverse correlation between miR-484 and ZFAS1 in CRC samples (E; p<0.05). Furthermore, we found that miR-484 expression was upregulated after ZFAS1 inhibited in CRC cells (F; p<0.05). RNA immunoprecipitation experiments showed that both miR-484 and ZFAS1 were in the Ago2-pulled down pellet (G; p<0.05). Thus, those data indicated that ZFAS1 might acted as a molecular sponge for miR-484.
Figure 4. LncRNA ZFAS1 directly interacted with miR-507 in CRC. (A) The predicted complementary sequence between miR-484 and ZFAS1 3’-UTR. (B) HEK293T cells were transfected with ZFAS1 3′-UTR Wt or Mut reporter plasmids along with miR-484 mimics, and then luciferase activity assays were performed. (C) Relative expression of miR-484 was measured by qRT-PCR in CRC tissues. (D) Relative expression of miR-484 was measured by qRT-PCR in CRC cells. (E) Pearson's correlation analysis between miR-484 and ZFAS1 expression in CRC tissues. (F) miR-484 expression in ZFAS1 knockdown CRC cells was determined by qRT-PCR. (G) Association of ZFAS1 and miR-484 with Ago2 in CRC cells. *p<0.05
miR-484 reversed the effect of ZFAS1 on CRC progression
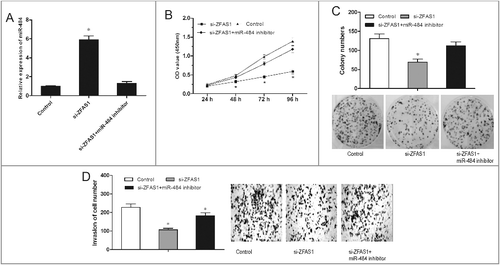
To explore whether ZFAS1 exerted biological functions through miR-484, we performed rescue experiments by inhibiting miR-484 expression in ZFAS1 knockdown SW480 cells (A; p<0.05). MTT and colony formation assay showed that miR-484 inhibitor could reverse the inhibition of ZFAS1 on proliferation capacity of SW480 cells (B and C; p<0.05). Transwell invasion assay showed that miR-484 inhibitor increased the invasion ability of SW480 cells reduced by ZFAS1 knockdown (D; p<0.05). Overall, miR-484 inhibitor could reverse the tumor suppressing of ZFAS1 knockdown on CRC cells, indicating the antagonism regulation of ZFAS1 and miR-484.
Figure 5. miR-484 reversed the effect of ZFAS1 on CRC tumorigenicity. (A) Relative expressions of miR-484 in CRC cells transfected with si-ZFAS1 and miR-484 inhibitor. (B) CCK-8 assay showed that proliferation of CRC cells transfected with si-ZFAS1, si-ZFAS1+miR-484 inhibitor. (C) Colony formation assay showed the clone number of CRC cells transfected with si-ZFAS1, si-ZFAS1+miR-484 inhibitor. (D) Transwell invasion assay showed the invasion of CRC cells transfected with si-ZFAS1, si-ZFAS1+miR-484 inhibitor. *p<0.05
Discussion
Despite the great therapeutic advances made in CRC, including surgical resection and adjuvant therapy, the long-term prognosis of CRC patients with distant metastases remains unfavorable [Citation18]. Increasing functional studies have indicated that lncRNAs, which may act as oncogenes or tumor suppressors, are involved in the carcinogenesis and progression of several tumors, including CRC [Citation19]. For example, Liu et al showed that enhanced expression of lncRNA Sox2ot promoted cell proliferation and motility in colorectal cancer [Citation20]. Wang et al revealed that lncRNA AB073614 regulated proliferation and metastasis of colorectal cancer cells via the PI3K/AKT signaling pathway [Citation21]. Liu et al suggested that lncRNA SPRY4-IT1 promoted proliferation and invasion by acting as a ceRNA of miR-101-3p in colorectal cancer cells [Citation22].
In the present study, we focused on the lncRNA ZFAS1 and found that its expression was significantly upregulated in CRC tissues and correlated with Helicobacter pylori infection, lymph nodes metastasis, advanced TNM stage, and poor overall survival. Furthermore, function experiments showed that ZFAS1 inhibition markedly suppressed the proliferation and invasion ability of CRC cells. In addition, our results showed that ZFAS1 knockdown suppressed tumor growth in vivo. Thus, our data strongly suggested that ZFAS1 could function as an oncogene in CRC tumorigenesis.
LncRNA is type of non-protein coding RNA and modulate pathophysiology process involving multi-levels regulation [Citation5]. One of the major regulating approach is to bind target miRNAs to absorb the miRNAs abundance and further modulate functional genes expression [Citation17]. For example, Zhang et al lncRNA UCA1 promoted cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer [Citation23]. Wang et al indicated that lncRNA TUG1 promoted migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells [Citation24].
Recent studies showed that miR-484 play important roles in tumor progression. For example, Hu et al showed that miR-484 suppressed proliferation and epithelial-mesenchymal transition by targeting ZEB1 and SMAD2 in cervical cancer cells [Citation25]. While, Li et al showed that miR-484 promoted non-small-cell lung cancer progression through inhibiting Apaf-1 associated with the suppression of apoptosis [Citation26]. However, the expression and roles of miR-484 on CRC remains unclear.
In the present study, qRT-PCR showed that miR-484 expression was significantly decreased and negatively correlated with ZFAS1 expression in CRC tissues. Bioinformatics prediction and luciferase reporter assay validated that miR-484 was the target of ZFAS1 sharing complementary binding sites at 3′-UTR. Besides, miR-484 reversed the functions of ZFAS1 on CRC cells proliferation and invasion, suggesting the competing endogenous RNA (ceRNA) mechanism in CRC progression.
In conclusion, our work revealed the overexpression of ZFAS1 and investigated the potential interaction with miR-484 in CRC tumorigenesis, indicating the ceRNA mechanism or miRNAs sponge in CRC. Thus, it could be argued the ZFAS1-miR-484 axis may become a new epigenetic therapeutic target for the treatment of CRC.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this Article.
Additional information
Funding
References
- Jemal A, Bray F, Center M M, et al. Global cancer statistics [J]. CA Cancer J Clin 2011;61(2):69–90. PMID:21296855
- Siegel R, DeSantis C, Jemal A. Colorectal cancer statistics, 2014 [J]. CA Cancer J Clin, 2014;64(2):104–117. PMID:24639052
- Aran V, Victorino AP, Thuler LC, et al. Colorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortality [J]. Clin Colorectal Cancer. 2016;15(3):195–203. doi:10.1016/j.clcc.2016.02.008. PMID:26964802
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development [J]. Nature Rev Gen. 2014;15(1):7. doi:10.1038/nrg3606. PMID:24296535
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions [J]. Nature Rev Gen. 2009;10(3):155. doi:10.1038/nrg2521. PMID:19188922
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas [J]. Mol Cancer. 2011;10(1):38. doi:10.1186/1476-4598-10-38. PMID:21489289
- Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases [J]. Cancer Lett. 2013;339(2):159–166. doi:10.1016/j.canlet.2013.06.013. PMID:23791884
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function [J]. Nature Rev Gen. 2016;17(1):47. doi:10.1038/nrg.2015.10. PMID:26666209
- Lin L, Gu ZT, Chen WH, et al. Increased expression of the long non-coding RNA ANRIL promotes lung cancer cell metastasis and correlates with poor prognosis [J]. Diagnostic Pathol. 2015;10(1):14. doi:10.1186/s13000-015-0247-7. PMID:25889788
- Li Z, Zhao L, Wang Q. Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/尾-catenin pathway [J]. Am J Trans Res. 2016;8(5):2385. PMID:27347346
- Wang Z, Yuan J, Li L, et al. Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137 [J]. Am J Trans Res. 2017;9(4):1845. PMID:28469789
- Zhang Y, Sun L, Xuan L, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction [J]. Scientific Reports. 2016;6. PMID:28442741
- Gao K, Ji Z, She K, et al. Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and promotes glioma cell progression by activation of the notch signaling pathway [J]. Biomed Pharmacotherapy. 2017;87:555–560. doi:10.1016/j.biopha.2017.01.014.
- Xia B, Hou Y, Chen H, et al. Long non-coding RNA ZFAS1 interacts with miR-150-5p to regulate Sp1 expression and ovarian cancer cell malignancy [J]. Oncotarget. 2017;8(12):19534. PMID:28099946
- Wang W, Xing C. Upregulation of long noncoding RNA ZFAS1 predicts poor prognosis and prompts invasion and metastasis in colorectal cancer [J]. Pathol Res Practice. 2016;212(8):690–695. doi:10.1016/j.prp.2016.05.003.
- Thorenoor N, Faltejskova-Vychytilova P, Hombach S, et al. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer [J]. Oncotarget. 2016;7(1):622. doi:10.18632/oncotarget.5807. PMID:26506418
- Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA [J]. Cell. 2011;147(2):358–369. doi:10.1016/j.cell.2011.09.028. PMID:22000014
- Van Cutsem E, Cervantes A, Nordlinger B, et al. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up [J]. Ann Oncol, 2014;25(suppl_3):iii1–iii9. doi:10.1093/annonc/mdu260. PMID:25190710
- Han D, Wang M, Ma N, et al. Long noncoding RNAs: novel players in colorectal cancer [J]. Cancer Lett. 2015;361(1):13–21. doi:10.1016/j.canlet.2015.03.002. PMID:25754818
- Liu S, Xu B, Yan D. Enhanced expression of long non-coding RNA Sox2ot promoted cell proliferation and motility in colorectal cancer [J]. Minerva Med. 2016;107(5):279–286. PMID:27353770
- Wang Y, Kuang H, Xue J, et al. LncRNA AB073614 regulated proliferation and metastasis of colorectal cancer cells via the PI3K/AKT signaling pathway [J]. Biomed Pharmacotherapy. 2017;93:1230–1237. doi:10.1016/j.biopha.2017.07.024.
- Liu D, Li Y, Luo G, et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2 [J]. Cancer Lett. 2017;388:281–291. doi:10.1016/j.canlet.2016.12.005. PMID:27998761
- Zhang S, Dong X, Ji T, et al. Long non-coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer [J]. Am J Trans Res. 2017;9(2):366. PMID:28337266
- Wang Y, Yang T, Zhang Z, et al. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells [J]. Cancer Sci. 2017. doi:10.1111/cas.13201.
- Hu Y, Xie H, Liu Y, et al. miR-484 suppresses proliferation and epithelial–mesenchymal transition by targeting ZEB1 and SMAD2 in cervical cancer cells [J]. Cancer Cell Int. 2017;17(1):36. doi:10.1186/s12935-017-0407-9. PMID:28286418
- Li T, Ding Z L, Zheng Y L, et al. MiR-484 promotes non-small-cell lung cancer (NSCLC) progression through inhibiting Apaf-1 associated with the suppression of apoptosis [J]. Biomed Pharmacotherapy. 2017;96:153–164. doi:10.1016/j.biopha.2017.09.102.
