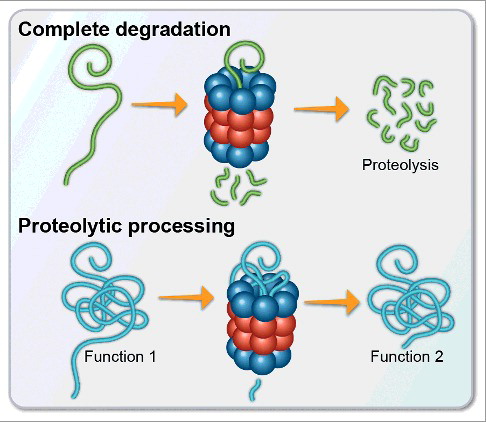
Protein degradation is an essential cellular process contributing to the maintenance of proteostasis. The removal of damaged and surplus proteins can be achieved by several means, predominantly involving proteasome complexes. The ubiquitin-dependent 26S proteasome is considered to be the major route that mediates this degradation. The 26S proteasome complex is comprised of the 19S regulatory caps, which are responsible for recognizing and unfolding ubiquitinated substrates, and a 20S catalytic core, which breaks peptide bonds within the substrate, leading to degradation. The 20S proteasome core can also exist independently of the 19S caps, and is capable of degrading substrates in an ubiquitin and ATP-independent manner (reviewed in 1). Unlike the substrates for the 26S proteasome which have been selectively modified with poly-ubiquitin tags, the substrates for the 20S proteasome are proteins that contain unstructured regions, either as a result of mutation or damage, or as an intrinsic feature of the proteins themselves, as is the case for intrinsically disordered proteins (IDPs) (A). Many key regulatory and signaling proteins, such as those involved in cell cycle control and transcription are IDPs, and it was indeed shown that the 20S proteasome plays a major role in the flux of their cellular levels [Citation1]. Recently, we and others have demonstrated that the activity of 20S proteasome is not restricted to complete degradation of its protein substrates, but rather there are proteins that the 20S proteasome cleaves at specific sites to generate functional cleavage products (B). This process influences diverse cellular pathways such as transcription, protein synthesis and the response to cellular stress [Citation2–6].
Figure 1. The 20S proteasome is capable of cleaving substrates leading to either full proteolysis of the unfolded substrate into small peptides, or to selective proteolysis of a specific disordered region within the polypeptide chain. The latter may lead to the generation of a degradation product that retains structural elements and has a differential functional property.
For example, subunits of the translation initiation factors eIF4F and eIF3 were shown to be endoproteolytically cleaved by the 20S proteasome [Citation2]. eIF4F and eIF3 are protein complexes involved in the recruitment of the 40S ribosome to mRNAs, thus allowing translation to proceed. The proteolytic processing by the 20S proteasome of eIF4G, a subunit of eIF4F, and eIF3a, a subunit of eIF3, was shown to lead to inhibition of the assembly of the pre-initiation complex and consequent prevention of the translation of certain mRNAs. In addition, the key NF-κB transcription factor family member, p50, was shown to be produced by endoproteolytic cleavage of its precursor, p105, by the 20S proteasome [Citation3]. p105 exists as a dimer, with the C-terminus of the protein folded such that the 20S proteasome processing region is exposed permitting cleavage, while a glycine rich region was identified as the stop signal for 20S proteasomal processing, protecting the N-terminal domains and allowing generation of the functional p50 product. This finding overturned the previously held model of p50 generation, which hypothesized that p50 was a product of co-translation caused by ribosomal pauses during translation of p105, and showed that p50 production occurs post-translationally and critically depends on 20S proteasomal processing [Citation3]. Hsp70, the central chaperone involved in the cellular stress response, is known to be a substrate of the 26S proteasome following recovery from stress. However, recent evidence revealed that Hsp70 can also undergo proteolytic cleavage by the 20S proteasome, leading to removal of the disordered C-terminus and generation of a 30 kDa product [Citation4]. While no function for this cleavage product was reported yet, this specific proteolytic processing of such an important chaperone by the 20S proteasome could have broad implications on our understanding of chaperone biology. Recently, we demonstrated that the 20S proteasome also cleaves the tumor suppressor p53 so as to coordinate its function [Citation5]. Cleavage by the 20S proteasome occurs precisely at position 40, generating the known Δ40p53 isoform. Thus, in addition to alternative translation initiation at position 40, Δ40p53 is generated by a post-translational event via the 20S proteasome. The Δ40p53 isoform is then capable of hetero-tetramerizing with the full length p53, attenuating downstream transcriptional activities. We showed that this negative feedback mechanism is particularly pronounced under conditions of oxidative stress, when the common MDM2-mediated regulation of p53 is disrupted while 20S proteasome activity is increased [Citation5].
The ability of the 20S proteasome to selectively degrade specific regions of its substrate proteins, leading to the modulation of protein function and alteration of downstream processes, is an exciting phenomenon that may have broad implications for cell function and survival. The generation of degradation products that retain structural and functional characteristics, as described in the previous examples, clearly demonstrate that the 20S proteasome plays a critical role beyond the substrate-to-peptides model for protein degradation. In addition, these studies have highlighted that substrate recognition by the 20S proteasome is not just limited to unfolded regions within the polypeptide chain, but is also influenced by the primary sequence, as demonstrated by the identification of the glycine rich region as a stop signal for proteasomal degradation [Citation3]. Considering that: i) the majority of proteasomes within the cell are 20S proteasomes [Citation7], ii) more than 20% of all cellular proteins are cleaved by the 20S [Citation6], and iii) close to half of the human proteome is predicted to contain intrinsically disordered segments (>30 amino acids in length), making them susceptible to degradation by the 20S proteasome (reviewed in 1), it is highly likely that future research will uncover even more widespread roles for the 20S proteasome. However, untangling the multifaceted roles of the 20S proteasome is complicated by the difficulties in distinguishing between 26S and 20S proteasome mediated degradation in vivo, as well as by the degradation of substrates by both proteasomal pathways. Further research into this aspect of 20S proteasomal proteolytic processing will shed light on regulatory principles of this process, the repertoire of substrates and beyond.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Ben-Nissan G, Sharon M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules. 2014;4:862–884. doi:10.3390/biom4030862. PMID:25250704.
- Baugh JM, Pilipenko EV. 20S proteasome differentially alters translation of different mRNAs via the cleavage of eIF4F and eIF3. Mol Cell. 2004;16:575–586. doi:10.1016/j.molcel.2004.10.017. PMID:15546617.
- Moorthy AK, Savinova OV, Ho JQ, et al. The 20S proteasome processes NF-kappaB1 p105 into p50 in a translation-independent manner. EMBO J. 2006;25:1945–1956. doi:10.1038/sj.emboj.7601081. PMID:16619030.
- Morozov AV, Astakhova TM, Garbuz DG, et al. Interplay between recombinant Hsp70 and proteasomes: proteasome activity modulation and ubiquitin-independent cleavage of Hsp70. Cell Stress Chaperon. 2017;22:687–697. doi:10.1007/s12192-017-0792-y. PMID:28447215.
- Solomon H, Bräuning B, Fainer I, et al. Post-translational regulation of p53 function through 20S proteasome-mediated cleavage. Cell Death Differ. 2017;24(12):2187–2198. doi:10.1038/cdd.2017.139. PMID:28885617.
- Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of Ubiquitination. J Mol Biol. 2009;386:814–827. doi:10.1016/j.jmb.2008.12.081. PMID:19162040.
- Fabre B, Lambour T, Garrigues L, et al. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J Proteome Res. 2014;13:3027–3037. doi:10.1021/pr500193k. PMID:24804812.
