ABSTRACT
Increasing evidence has shown that abnormal expression of lncRNAs is involved in various biological behaviors and major cellular pathways of human cancers. However, the role of lncRNAs in the progression of gastric cancer has not been adequately investigated. Therefore, in this study, we investigated the expression levels of linc-GPR65-1 using Quantitative real-time PCR (qRT-PCR) and found that linc-GPR65-1 was significantly up-regulated in 50 gastric cancer tissues compared to the corresponding normal tissues. In addition, increased linc-GPR65-1 expression was associated with TNM stage (P = 0.037), tumor size (P = 0.024), distal metastasis (P = 0.023), and poor prognosis of gastric cancer patients. Moreover, functional assays indicated that decreased linc-GPR65-1 expression inhibited the aggressive phenotypes of gastric cancer cells, and enhanced linc-GPR65-1 expression resulted in the opposite phenomenon. Then, a cancer signaling phosphoantibody microarray was conducted to explore the potential mechanisms of linc-GPR65-1 in regulating gastric cancer progression and observed that linc-GPR65-1 could regulate the PTEN-AKT-slug signaling pathway. These data showed that linc-GPR65-1, regulating the PTEN-AKT-slug signaling pathway, might act as a tumor promoter and serve as a novel target for gastric cancer prevention and therapy.
Introduction
The incidence and mortality of gastric cancer have slightly decreased in recent years, but this malignancy remains a serious threat to human health worldwide [Citation1,Citation2]. Although gastric cancer is curable in the early stages, the majority of gastric cancer patients are diagnosed in advanced stages and tend to have an unfavorable prognosis [Citation3]. Various abnormal molecular changes have been identified in gastric cancer [Citation4,Citation5]. However, the underlying molecular mechanisms regulating the aggressive phenotypes of gastric cancer remain unclear [Citation6]. Therefore, elucidation of the molecular mechanisms and development of improved therapeutic strategies for gastric cancer are urgently needed.
Recently, long non-coding RNAs (lncRNA), which are longer than 200 nucleotides in length, have attracted the attention of many researchers. Increasing evidence has shown that abnormal expression of lncRNAs is involved in various biological behaviors and major cellular pathways of human cancers, such as proliferation, invasion, migration, the cell cycle and apoptosis [Citation7-9]. In addition, recent data have shown that several lncRNAs exhibit distinct gene expression patterns in gastric cancer [Citation10,Citation11]. However, the role of lncRNAs in the progression of gastric cancer has not been adequately investigated.
To explore the roles of lncRNAs in gastric cancer, we performed transcriptome sequencing of four pairs of gastric cancer and normal tissues (Annoroad Technology, Beijing, China). Among the differentially expressed mRNAs and lncRNAs, linc-GPR65-1 was significantly up-regulated in gastric cancer tissues compared to the corresponding normal tissues. This long intergenic non-protein coding RNA, which is located on chromosome 14, is comprised of 413 nucleotides. The expression, biological functions and potential molecular mechanisms of linc-GPR65-1 in regulating gastric cancer have not been investigated Therefore, in the present study, we examined the expression, biological functions and molecular mechanisms of linc-GPR65-1 in gastric cancer. First, the expression levels of linc-GPR65-1 were detected in 50 pairs of gastric cancer tissues, and its clinical value was analyzed. Then, we investigated the effect of linc-GPR65-1 on biological phenotypes of gastric cancer cells. Finally, a cancer signaling phosphoantibody microarray was performed to elucidate the potential mechanisms of linc-GPR65-1 in regulating gastric cancer progression. Thus, we identified a novel lncRNA as a potential therapeutic target for gastric cancer.
Results
Linc-GPR65-1 is up-regulated in gastric cancer tissues and has clinical value in gastric cancer
The expression of linc-GPR65-1 in 50 gastric cancer tissues and paired normal tissues was detected using qRT-PCR, and our results showed that linc-GPR65-1 was significantly up-regulated in gastric cancer tissues (A). Furthermore, based on the qRT-PCR data, all 50 patients were divided equally into two groups according to the median expression level of linc-GPR65-1 (B). In addition, we analyzed the clinical characteristics associated with linc-GPR65-1 expression. As shown in , the linc-GPR65-1 level was significantly associated with tumor size (P = 0.024), TNM stage (P = 0.037) and distant metastasis (P = 0.023). A significant correlation with other features, such as age, gender, differentiation, and lymph node metastasis, was not observed. Moreover, we also analyzed the clinical value of linc-GPR65-1 in predicting the prognosis of gastric cancer patients using Kaplan–Meier analysis, and the results indicated that high linc-GPR65-1 expression was associated with poor survival rate (P = 0.037; C).
Figure 1. Expression of linc-GPR65-1 in gastric cancer tissues and its clinical value in overall survival. A. The expression of linc-GPR65-1 was up-regulated in gastric cancer tissues compared to adjacent normal tissue (P < 0.01). B. All 50 gastric cancer samples were divided into two groups, with 25 patients in each group, according to the median expression level. C. Kaplan–Meier analysis showed that a high level of linc-GPR65-1 indicated a worse prognosis of gastric cancer patients (P = 0.037).
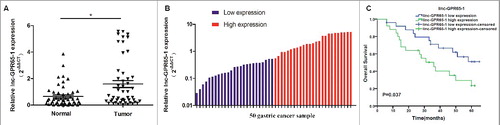
Table 1. Association between clinicopathological characteristics and linc-GPR65-1 expression
Expression of linc-GPR65-1 in gastric cancer cell lines and its subcellular localization
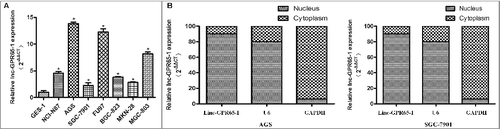
Then, the expression levels of linc-GPR65-1 were estimated in 7 gastric cancer cell lines and normal gastric mucosal epithelial cells (GES-1). As shown in A, the expression of linc-GPR65-1 was higher in all 7 gastric cancer cell lines than the GES-1 cell line, with the highest levels in AGS cells and lowest in SGC-7901 cells; thus, these lines were used for further analyses of the biological functions and mechanisms. Moreover, the subcellular localization of linc-GPR65-1 was determined in AGS and SGC-7901 cells using nucleus-cytoplasm fractionation, as shown in B. We observed that linc-GPR65-1 was primarily expressed in the nucleus, which suggested that linc-GPR65-1 may play an important role in altering cell functions at the transcriptional level or bind to nucleoproteins.
Silencing of linc-GPR65-1 expression in gastric cancer cells inhibits cell proliferation, migration and invasion in vitro
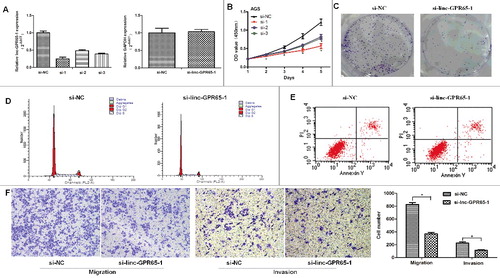
Next, to elucidate the biological functions of linc-GPR65-1 in gastric cancer, we knocked down the expression of linc-GPR65-1 using siRNA or a negative control in AGS cells. The expression of linc-GPR65-1 was down-regulated approximately 75%, 50%, 60% after treated with three different siRNA targeting linc-GPR65-1 compared to the negative control. And, GAPDH is not changing upon lncRNA knockdown when normalized to β-actin (A). Silenced linc-GPR65-1 expression significantly inhibited cell proliferation, as evaluated by CCK-8 assays (B), and inhibited colony formation ability (C). AGS cells treated with si-linc-GPR65-1 exhibited cell cycle arrest compared with si-NC cells (G0/G1%: 73.71% vs. 67.20%, P < 0.01) (D), but there was no effect on apoptosis rate (11.94% vs. 11.25%, P = 0.337) (E), which was detected using flow cytometry analysis. Finally, cell migration and invasion were evaluated by transwell assays, and increased expression of lnc-GPR65-1 significantly inhibited the migration and invasion of AGS cells (F).
Figure 3. Decreased linc-GPR65-1 inhibited proliferation, cell motility, invasion and migration of AGS cells. A. The knockdown efficiency of siRNA against linc-GPR65-1 was confirmed using qRT-PCR, and the efficiency was approximately 75%, 50%, 60% after treated with three different siRNA, respectively. And GAPDH is not changing upon lncRNA knockdown when using another internal reference (β-actin). B. Cell proliferation was detected via CCK-8 assays in the si-linc-GPR65-1 group and the negative control group during a period of 5 days. C. Colony formation assays were also performed to estimate the cell proliferation capacity. D.E. Cell cycle and apoptosis rate of AGS cells were evaluated by flow cytometry. F. The invasion (right) and migration (left) of AGS cells were evaluated in the si-linc-GPR65-1 group and the negative control group by transwell assays (magnification × 100).
Enhanced linc-GPR65-1 expression in gastric cancer cells promotes cell proliferation, migration and invasion in vitro
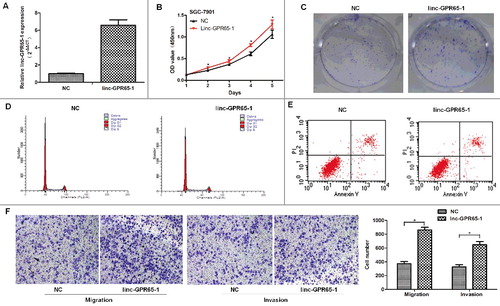
To further investigate the role of linc-GPR65-1 in gastric cancer, we enhanced the expression of linc-GPR65-1 in SGC-7901 cells using the pcDNA3.1 plasmid vector. The expression of linc-GPR65-1 was up-regulated approximately 7 times after treated with pcDNA3.1 plasmid vector compared to the negative control (A). The CCK-8 and colony formation assays showed that the proliferation of SGC-7901 cells increased after treatment with the linc-GPR65-1 plasmid compared with the negative control (B. C). SGC-7901 cells treated with the linc-GPR65-1 plasmid showed a significantly arrested cell cycle compared with NC cells (G0/G1%: 67.06% vs. 70.41%, P < 0.01) (D), but there was no significant difference between these two groups in the apoptosis rate (15.34% vs. 15.98%, P = 0.202) (E). Transwell assays showed that enhanced expression of linc-GPR65-1 promoted the migration and invasion of SGC-7901 cells (F).
Figure 4. Increased linc-GPR65-1 promoted proliferation, cell motility, invasion and migration of SGC-7901 cells. A. The over-expression efficiency after treatment with linc-GPR65-1 plasmid; the 7-fold change was verified using qRT-PCR. B. Cell proliferation was detected by CCK-8 assays in the linc-GPR65-1 over-expression group and the negative control group. C. Colony formation assays were also performed to evaluate the cell proliferation capacity. D.E. Flow cytometry was used to detect the cell cycle and apoptosis rate of SGC-7901 cells. F. The migration (left) and invasion (right) of SGC-7901 cells were evaluated in the linc-GPR65-1 over-expression group and the negative control group by transwell assays (magnification × 100).
The above experiments performed with siRNA or an over-expression plasmid suggested that increased linc-GPR65-1 expression promotes gastric cancer tumorigenicity, which was consistent with the clinical findings.
Linc-GPR65-1 acts as a tumor promoter through the PTEN-AKT-slug pathway
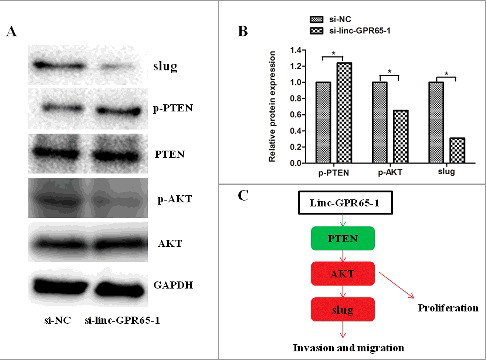
To comprehensively elucidate the mechanisms of linc-GPR65-1 in regulating gastric cancer cell proliferation, migration and invasion, we performed a phosphoprotein antibody array. AGS cells treated with si-linc-GPR65-1 lentiviral vector or negative control were analyzed to identify potential proteins regulated by linc-GPR65-1. A total of 47 proteins were shown to be altered by more than 12% after down-regulation of linc-GPR65-1; various proteins were enriched in the PI3K signaling pathway and play important roles in regulating tumor development (Supplemental Table 1). We confirmed using western blot analysis that PTEN phosphorylation was increased and phosphorylation of AKT was significantly reduced following treatment with si-linc-GPR65-1 compared to si-NC ().
Discussion
Gastric cancer poses a major public health threat worldwide. Although gastric cancer has been intensively studied, the underlying molecular mechanisms have not been fully elucidated. In addition, there are no effective prevention and treatment strategies for gastric cancer. Therefore, identification of effective predictors and treatments is urgently needed.
Several studies have indicated that the mechanism of carcinogenesis is not limited to protein-coding genes [Citation12], and thus, lncRNAs have been increasingly investigated by researchers. Based on our previous data (unpublished), we identified a novel lncRNA known as linc-GPR65-1. First, we detected the expression of linc-GPR65-1 and found that it was significantly up-regulated in gastric cancer tissues compared to the corresponding adjacent normal tissues. Then, clinical characteristics were analyzed, and the data indicated that increased lnc-GPR65-1 expression in gastric cancer tissues was significantly related to tumor size, TNM stage and distant metastasis. Based on the prognosis of patients with gastric cancer [Citation13], we assessed whether linc-GPR65-1 can act as a predictor of prognosis. Our data demonstrated that high expression of linc-GPR65-1 was associated with a poor prognosis. These findings indicated that this molecule has important research significance. Therefore, further studies on the role of linc-GPR65-1 were performed in this study.
To explore the biological functions of linc-GPR65-1, we silenced and enhanced its expression. Our experiments showed that decreased linc-GPR65-1 expression could inhibit the malignant biological behavior of gastric cancer cells, while increased linc-GPR65-1 expression had the opposite effect. Therefore, these results supported the hypothesis that linc-GPR65-1 could accelerate tumor development and also promoted cell proliferation and metastasis of gastric cancer. However, it was unclear how linc-GPR65-1 promotes tumor progression. Although the detailed mechanisms require further elucidate lncRNAs, which were predominantly located in the cell nucleus, play an important role in regulating the biological behavior of gastric cancer cells by interacting with proteins [Citation14]. In addition, in this study, we found that linc-GPR65-1 was predominantly located at the cell nucleus, which suggests it promotes gastric cancer progression by interaction with proteins. Previous studies have reported various molecular mechanisms; lnc-SNHG5 interacted with MTA2 [Citation15], lnc-HIT interacted with E2F1 [Citation16] and lnc-MALAT1 interacted with EZH2 [Citation17]. Slug, a key mediator of EMT, was associated with an aggressive phenotype of gastric cancer and was shown to bind to linc00216 [Citation18]. Our invasion and migration assays indicated that linc-GPR65-1 may be associated with epithelial-mesenchymal transition (EMT). Thus, we also explored EMT factors and found that slug was significantly down-regulated after treatment with si-linc-GPR65-1, which indicates that it might be downstream of linc-GPR65-1; these results were confirmed using western blot analysis. Thus, our findings indicated that linc-GPR65-1 promotes gastric cancer cell proliferation and metastasis by activating slug expression. However, the molecular mechanisms of linc-GPR65-1 in regulating slug or the signaling pathways involved in both lincGPR65-1 and slug remain unclear. Recently, several studies demonstrated that lncRNAs could regulate the expression of downstream genes through well-known signaling pathways. For example, the long non-coding RNA BANCR regulated liver cancer cell behavior through the MAPK signaling pathway [Citation19], the long non-coding RNA GNAT1-1 inhibited colorectal cancer cell proliferation and migration via the NF-κB signaling pathway [Citation20], and the long non-coding RNA HULC regulated cell proliferation of chronic myeloid leukemia through the PI3K/AKT signaling pathway [Citation21]. In addition, similar regulatory mechanisms can also be found in gastric cancer [Citation22-24]. These reports prompted us to perform further studies. Therefore, a cancer signaling phosphoantibody microarray was conducted to explore potential proteins that were altered in gastric cancer cells after treatment with the si-linc-GPR65-1 lentiviral vector compared with the negative control. The results showed that all 47 proteins, including phospho-PTEN and phospho-AKT, were significantly altered in the si-linc-GPR65-1 group compared with the negative control. In addition, KEGG pathway analysis showed that these proteins were enriched in the PI3K-AKT signaling pathway, which was consistent with our findings. Therefore, we examined the PI3K-AKT signaling pathway and confirmed the results using western blot. The PI3K-AKT molecular pathway, which can promote cell survival and cell proliferation, has been extensively researched in numerous types of tumors [Citation25,Citation26] and has been shown to genetically explain most cases of gastric cancer [Citation27-30]. These data indicated that linc-GPR65-1 is a tumor-promoting gene that regulates the PTEN-AKT signaling pathway and slug expression in gastric cancer. Therefore, linc-GPR65-1 may promotes gastric cancer cell growth and metastasis via the PTEN-AKT-slug signaling pathway.
In conclusion, our data indicated that linc-GPR65-1 expression was significantly up-regulated in gastric cancer tissues and cells. Linc-GPR65-1 was shown to possess clinical value, as it was a predictor of prognosis in patients with gastric cancer. Furthermore, linc-GPR65-1 was involved in promoting gastric cancer cell proliferation and metastasis, and its molecular mechanisms were elucidated. Therefore, linc-GPR65-1 may provide novel insights into developing molecular targeted therapies.
Materials and methods
Patients and tissue specimens
Frozen tissues of gastric cancer and normal tissues, which were collected from January 2010 to December 2010, were reserved in the biological samples library of Peking University People’s Hospital. Each patient provided informed consent, and collections were approved by the ethics committee. All patients were followed-up for 5 years and received no anti-tumor treatment before operation. For the use of tissues, each patient was informed and provided consent for research, and this study was approved by the ethics committee of the Beijing University People's Hospital.
Cell culture and transfection
GES-1, MKN-28, SGC-7901, HGC-27, NCI-N87, AGS, and BGC-823 cells were preserved in our laboratory and cultured in DMEM-F12 (1:1) medium. FU97 was purchased from JCRB and was cultured in DMEM with 10 mg/l insulin. The medium was supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, USA) at 37°C with 5% CO2. For transfection, 2 × 104 cells/well were pre-cultured in 12-well plates for 24 h and then infected using Lipofectamine 3000 (Invitrogen, Carlsbad, CA) according to the manufacturer's directions. The linc-GPR65-1 siRNA sequence: ENST00000553929-homo-154 (si-1): 5′-GCUAGAGGCAUCCUGCAAUTT-3′; ENST00000553929-homo-252 (si-2): 5′-GUUCUUCAGAUGAGUAGAATT-3′; ENST00000553929-homo-376 (si-3): 5′-GACCUCCAAACAAUCACCATT-3′. siRNAs were purchased from GenePharma (GenePharma Co., Ltd., Suzhou, China).
For in vitro functional assays, siRNAs targeting linc-GPR65-1 were designed and synthetized (GenePharma, Shanghai, China), and a final concentration of 50 nM was used; the full-length human linc-GPR65-1 cDNA was cloned into the pcDNA3.1 expression vector (GenePharma, Shanghai, China), and a final concentration of 2 µg/ml was used.
For the cancer signaling phosphoantibody microarray, the siRNA lentivirus vector LV3-GFP-Puro was constructed, and stable transfected cells were established.
RNA isolation and qRT-PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA) as described in the manufacturer's instructions. PrimeScript™ RT Master Mix (TaKaRa) was used for reverse transcription, and real-time PCR was performed using iTaq™ Universal SYBR Green Supermix (Bio-Rad, CA, USA). The primer sequences of linc-GPR65-1 are as follows: forward: 5′-AGCCCCACCTTCTGCTACTA-3′; reverse: 5′-TCTTCCTGCTCTCTCCCTCC-3′. All reactions were repeated 3 times. The fold change for each gene was calculated using the 2−ΔΔCt method, and gene expression was normalized to that of GAPDH.
Colony formation
For colony formation assays, a 1000 cell suspension of gastric cancer cells was plated into 6-well plates and cultured for 10 days. Colonies with at least 50 cells were considered significant. The experiments were performed three times, and the results are shown as the mean ± standard deviation.
Cell proliferation and cell cycle assays
For proliferation assay, 1000 single suspended gastric cancer cells with complete medium were cultured in 96-well plates for 0 h, 24 h, 48 h, 72 h, 96 h and 120 h. The cell number was estimated using a CCK-8 kit (Dojindo, Japan) according to the manufacturer's instruction. Each group was assayed six times.
For cell cycle analysis, cells were stained with PI solution (BD Cycletest™ Plus DNA Kit) and were analyzed using a fluorescence-activated cell sorter after a 24 h transfection.
Invasion and migration assays
For the migration assay, 4 × 104 cells in 200 µl medium with 3% FBS were added to the upper chamber, and 600 µl medium with 10% FBS was added to the lower chamber (Product #3422, Corning Costar Corp, Cambridge, MA, USA). Then, the membranes were stained with crystal violet following 24 h of culture.
For invasion assays, 8 × 104 cells were added to the upper chamber, and the culture conditions were identical to those of the migration assay but with 50 µl Matrigel on the membranes. Then, the membranes were stained with crystal violet following 48 h of culture.
Apoptosis assay
For apoptosis assays, 1 × 105 cells were collected after a 24 h transfection and stained with 5 µl FITC-Annexin V and 5 µl PI for 15 min using the Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA). Then, 400 µl 1 × binding buffer was added to the cells, which were analyzed using a FACScan flow cytometry system (BD Biosciences, San Jose, CA, USA) within 1 h. Data were analyzed using FlowJo V7 software (Tree Star, Ashland, OR, USA).
Western blot analysis
Total protein was extracted from AGS cells using RIPA buffer (PMSF/RIPA: 1:100), and protein concentration was determined by the BCA method. Then, 50 µg total proteins were separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (General Electric Healthcare, Buckinghamshire, UK). Membranes were incubated with primary antibodies against slug, AKT, Phospho-AKT (ser473), PTEN, and Phospho-PTEN (Ser380/Thr382/Thr383) (CST, Danvers, MA). All antibodies were diluted 1:1000. GAPDH was used as a control.
Cancer signaling phosphoantibody microarray
For further analysis of the potential mechanisms of how linc-GPR65-1 increased slug expression and affected gastric cancer cell proliferation, migration and invasion, a cancer signaling phosphoantibody microarray was performed in cooperation with Wayen Biotechnology (Shanghai, China) after cells were treated with si-linc-GPR65-1 and si-NC. This assay can rapidly detect 248 cancer signaling phosphoantibodies with three replicates. Finally, data were collected and analyzed based on their established protocol.
Statistical analysis
All data are expressed as the mean ± SD and were analyzed using SPSS 18.0. Differences between groups were assessed using the χ2 test, Student's t-test or Fisher’s exact test. The overall survival was analyzed by the Kaplan–Meier method. The median was used to define the threshold values of low or high expression of linc-GPR65-1. P < 0.05 was considered significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
2017CC7746R1-file003.docx
Download MS Word (18.6 KB)Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332. PMID:26742998
- Chen W, Zheng R, Baade PD, et al. Cancer Statistics in China, 2015. CA Cancer J Clin. 2016;66(2): 115–132. doi:10.3322/caac.21338. PMID:26808342
- Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi:10.3748/wjg.v12.i1.17. PMID:16440411
- Guo XB, Jing CQ, Li LP, et al. Down-regulation of miR-622 in gastric cancer promotes cellular invasion and tumor metastasis by targeting ING1 gene. World J Gastroenterol. 2011;17:1895–1902. doi:10.3748/wjg.v17.i14.1895. PMID:21528065
- Xu TP, Liu XX, Xia R, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45): 5648–5661. doi:10.1038/onc.2015.18. PMID:25728677
- Milne AN, Carneiro F, O'Morain C, et al. Nature meets nurture: molecular genetics of gastric cancer. Hum Genet. 2009;126(5):615–628. doi:10.1007/s00439-009-0722-x. PMID:19657673
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi:10.1016/j.cell.2009.02.006. PMID:19239885
- Li JK, Chen C, Liu JY, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017;16(1):111. doi:10.1186/s12943-017-0681-0. PMID:28659173
- Jiang X, Liu W. Long Noncoding RNA highly upregulated in liver cancer activates p53-p21 pathway and promotes nasopharyngeal carcinoma cell growth. DNA Cell Biol. 2017;36(7): 596–602. doi:10.1089/dna.2017.3686. PMID:28445086
- Bi M, Yu H, Huang B, et al. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in gastric cancer cells through regulating CDKN1A. Gene. 2017;626:337–343.
- Yan K, Tian J, Shi W, et al. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42(3): 999–1012. doi:10.1016/j.gene.2017.05.049. doi:10.1159/000478682. PMID:28683446.. PMID:28571676.
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi:10.1146/annurev-biochem-051410-092902. PMID:22663078
- Li YS, Jing CQ, Chen YZ, et al. Expression of tumor necrosis factor α-induced protein 8 is upregulated in human gastric cancer and regulates cell proliferation, invasion and migration. Mol Med Rep. 2015;12(2): 2636–2642. doi:10.3892/mmr.2015.3690. PMID:25936980
- Huang M, Hou J, Wang Y, et al. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol Ther. 2017;25(4): 1014–1026. doi:10.1016/j.ymthe.2017.01.017. PMID:28214253
- Zhao L, Guo H, Zhou B, et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene. 2016;35(44): 5770–5780. doi:10.1038/onc.2016.110. PMID:27065326
- Yu L, Fang F, Lu S, et al. lncRNA-HIT promotes cell proliferation of non-small cell lung cancer by association with E2F1. Cancer Gene Ther. 2017;24(5): 221–226. doi:10.1038/cgt.2017.10. PMID:28429752
- Hirata H, Hinoda Y, Shahryari V, et al. Long Noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75(7): 1322–1331. doi:10.1158/0008-5472.CAN-14-2931. PMID:25600645
- Yu Y, Li L, Zheng Z, et al. Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slugdegradation. J Cell Mol Med. 2017;21(5): 955–967. doi:10.1111/jcmm.13035. PMID:27878953
- Gan Y, Han N, He X, et al. Long non-coding RNA CASC2 regulates cell biological behaviour through the MAPK signaling pathway in hepatocellular carcinoma. Tumour Biol. 2017;39(6):1010428317706229. doi:10.1177/1010428317706229. PMID:28621238
- Ye C, Shen Z, Wang B, et al. A novel long non-coding RNA lnc-GNAT1-1 is low expressed in colorectal cancer and acts as a tumor suppressor through regulating RKIP-NF-κB-Snail circuit. J Exp Clin Cancer Res. 2016;35(1): 187. doi:10.1186/s13046-016-0467-z. PMID:27912775
- Lu Y, Li Y, Chai X, et al. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathwayin chronic myeloid leukemia. Gene. 2017;607:41–46. doi:10.1016/j.gene.2017.01.004. PMID:28069548
- Ji J, Tang J, Deng L, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6(40): 42813–42824. doi:10.18632/oncotarget.5970. PMID:26540343
- Zhang ZX, Liu ZQ, Jiang B, et al. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-κB1. Biochem Biophys Res Commun. 2015;465(2): 225–231. doi:10.1016/j.bbrc.2015.07.158. PMID:26248136
- Li P, Xue WJ, Feng Y, et al. Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am J Transl Res. 2016;8(8): 3522–3529.
- Spangle JM, Roberts TM, Zhao JJ. The emerging role of PI3K/AKT-mediated epigenetic regulation in cancer. Biochim Biophys Acta. 2017;1868(1): 123–131. PMID:28315368.. PMID:27648142.
- Lin A, Piao HL, Zhuang L, et al. FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacologic inhibition of the PI3K-AKT pathway. Cancer Res. 2014;74:1682–1693.
- Nasser MI, Masood M, Wei W, et al. Cordycepin induces apoptosis in SGC-7901 cells through mitochondrial extrinsic phosphorylation ofPI3K/Akt by generating ROS. Int J Oncol. 2017;50(3):911–919. doi:10.1158/0008-5472.CAN-13-1729. doi:10.3892/ijo.2017.3862. PMID:28197639. PMID:24448243.
- Wang D, Xin Y, Tian Y, et al. Pseudolaric acid B inhibits gastric cancer cell metastasis in vitro and in haematogenous dissemination model through PI3K/AKT, ERK1/2 and mitochondria-mediated apoptosis pathways. Exp Cell Res. 2017;352(1): 34–44. doi:10.1016/j.yexcr.2017.01.012. PMID:28132880
- Wei S, Wang L, Zhang L, et al. ZNF143 enhances metastasis of gastric cancer by promoting the process of EMT throughPI3K/AKT signaling pathway. Tumour Biol. 2016;37(9): 12813–12821. doi:10.1007/s13277-016-5239-z. PMID:27449034
- Wang SQ, Wang C, Chang LM, et al. Geridonin and paclitaxel act synergistically to inhibit the proliferation of gastric cancer cells through ROS-mediated regulation of the PTEN/PI3K/Akt pathway. Oncotarget. 2016;7(45): 72990–73002. doi:10.18632/oncotarget.12166. PMID:27659528