ABSTRACT
To easily edit the genome of naïve human embryonic stem cells (hESC), we introduced a dual cassette encoding an inducible Cas9 into the AAVS1 site of naïve hESC (iCas9). The iCas9 line retained karyotypic stability, expression of pluripotency markers, differentiation potential, and stability in 5iLA and EPS pluripotency conditions. The iCas9 line induced efficient homology–directed repair (HDR) and non-homologous end joining (NHEJ) based mutations through CRISPR-Cas9 system. We utilized the iCas9 line to study the epigenetic regulator, PRC2 in early human pluripotency. The PRC2 requirement distinguishes between early pluripotency stages, however, what regulates PRC2 activity in these stages is not understood. We show reduced H3K27me3 and pluripotency markers in JARID2 2iL-I-F hESC mutants, indicating JARID2 requirement in maintenance of hESC 2iL-I-F state. These data suggest that JARID2 regulates PRC2 in 2iL-I-F state and the lack of PRC2 function in 5iLA state may be due to lack of sufficient JARID2 protein.
Introduction
hESC have the capacity to self-renew indefinitely and differentiate into all cell types of the body [Citation1]. These cells have high potential for increasing the basic understanding of human biology during early development, and facilitating medical regenerative therapies in the future. Both approaches require the capacity to readily manipulate the genome to identify the key regulators of the process. While low genome editing efficiency in hESC has been a problem previously, the emergence of CRISPR gene editing platforms now allows rapid and efficient genetic manipulations in these cells. Stem cell based disease modeling holds great potential to study human biological systems such as neurological diseases, where access to living tissue from affected patients is not usually possible [Citation2]. Patient derived iPSC are currently employed to differentiate into human disease-relevant cell types, however comparisons between lines from different individuals are complicated by genomic variation between individuals and limited patient tissues. Complementary to work in iPSC, introduction of the genetic variant of interest in hESC is another potential approach to in vitro disease modeling [Citation3–5]. Efficient gene editing to systematically introduce disease relevant genetic variants into one common “wildtype” line allows for direct comparison of variants on protein function or intracellular pathways within the same genetic background.
Methods for rapid and controllable genetic manipulations using the CRISPR platform have recently been developed for hESC in the primed stage of pluripotency [Citation5–8]. These new methods allow highly efficient generation of biallelic knockouts in primed hESC or iPSC populations. Several stages of pluripotency have been isolated and maintained in culture in mouse and human: naïve ESC correspond to pre-implantation embryonic ICM and primed correspond to post-implantation embryonic epiblast. Multiple naive, pre-implantation human pluripotent stages have been stabilized in vitro [Citation9–18] The human naïve state has been defined by growth characteristics, mRNA and microRNA expression, epigenetic profile, Oct4 enhancer usage, X-inactivation profile, mitochondrial morphology, metabolic profile and development in the context of teratomas and chimeras. These hESC can be passaged as single cells, allowing easy genetic manipulations and thereby better genetic modification capacity using a CRISPR-Cas9 system. Indeed, a recent study shows that the efficiency of CRISPR-based gene editing is higher in in vitro toggled, naïve hiPSCs compared to primed [Citation19]. It has been proposed that this may be due to DNA hypomethylation and lower H3K27me3 marks, allowing more open chromatin structure that is more accessible for Cas9.
We now report the generation of a rapid, multiplex and inducible gene editing system in naive, pre-implantation hESC (Elf1-iCas9). In this study, we engineered a naïve hESC line for efficient gene editing and knock-out platforms by inserting the inducible Cas9 gene into the safe-harbor locus AAVS1 in the naïve hESC Elf1. Using this naive hESC platform, we generated heterozygous and homozygous missense mutations in the PSEN2 gene associated with autosomal dominantly inherited familial Alzheimer's Disease. We also show highly efficient single guide NHEJ-based mutant generation for multiple genes including TCTN2, Mel18L, NNMT, HIF1α, HIF2α, IDO1, PKLR, GPI and JARID2. However, while indels are identified in over 90% of the clones, the JARID2 and HIF2α mutant clone analysis revealed that only 18% of these mutations cause total lack of protein (protein null mutations). We used this efficient iCas9 hESC line to study the function of JARID2 in early human pluripotency. The JARID2 2iL-I-F hESC mutants showed reduced H3K27me3 epigenetic repressive marks and reduced stem cell marker signature, showing that JARID2 is required to maintain the stem cell 2iL-I-F pluripotency state. PRC2 is not required in earliest naïve state in mouse or human but shows requirement in the 2iL-I-F state [Citation20]. We show that while other PRC2 components are expressed in both stages, JARID2 is dramatically downregulated in the earliest, 5iLA but not in the 2iL-I-F state. We have now revealed JARID2 requirement for the PRC2 activity in the 2iL-I-F state. These data suggest that lack of PRC2 function in the earliest, naïve 5iLA state may be due to highly reduced JARID2 protein.
Experimental procedures
Cell culture
Naïve hESC [Elf-1(NIH_hESC Registry #0156), and newly Seattle-derived Elm2, Elf3 and Elf4] were cultured as previously described [Citation12]. All 4 hESC lines have a normal, diploid karyotype. Elm2 is male, Elf1, Elf3 and Elf4 are females. For 2iL-I-F conditions the cells were grown on a feeder layer of irradiated primary mouse embryonic fibroblasts in hESC media: DMEM/F-12 media supplemented with 20% knock-out serum replacer (KSR), 0.1mM nonessential amino acids (NEAA), 1 mM sodium pyruvate, and penicillin/streptomycin (all from Invitrogen, Carlsbad, CA) and 0.1 mM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO). hESC media was supplemented with 1µM GSK3 inhibitor (CHIR99021, Selleckchem), 1µM of MEK inhibitor (PD0325901, Selleckchem), 10ng/mL human LIF (Chemicon), 5ng/mL IGF1 (Peprotech) and 10ng/mL bFGF. For 5iLA naïve conditions [Citation21] cells were grown in base medium containing: 120 ml DMEM/F12 (Invitrogen; 11320), 120 ml Neurobasal (Invitrogen; 21103), 2.5 ml N2 supplement (Invitrogen; 17502048), 5 ml B27 supplement (Invitrogen; 17504044), 1 mM glutamine (Invitrogen), 1% nonessential amino acids (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma), penicillin-streptomycin (Invitrogen), and 50 mg/ml BSA (Sigma), and freshly supplemented with 5i/L/A: BRAF (0.5µM), SRC (1µM), MEKi (1µM), GSK3i (1µM), ROCKi (10µM), recombinant human LIF (20ng/mL), and Activin A (10ng/mL). For EPS conditions (extended pluripotency conditions) [Citation18], cells were grown in base medium containing 100 mL DMEM/F12, 100 mL Neurobasal, 1 mL N2 supplement, 2 mL B27 supplement, 1% GlutaMAX, 1% NEAA, 0.1 mM β-mercaptoethanol, penicillin-streptomycin and 5% KSR, and freshly supplemented with 10 ng/ml hLIF, GSK3i (1 μM), ROCKi (2 μM), (S)-(+)-Dimethindene maleate (2 μM; Tocris), Minocycline hydrochloride (2 μM; Santa Cruz Biotechnology) and IWR-endo-1 (0.5–1 μM; Selleckchem). Cells were adapted to 5i/L/A or EPS conditions for at least 3 passages before analysis. Elf1-iCas9 cells were pushed toward differentiation using DMEM medium supplemented with 20% FBS and retinoic acid (0.1μM RA; Sigma) for 5 days. All cells were cultured at 37 degrees Celsius in 5% CO2. Elf1-iCas9 were generated in low (5%) O2 and further expanded in normoxia (20% O2) for experimental studies. Cells were exposed to 2% O2 for 4 hours prior to protein extraction for the analysis of HIF2α expression.
Insertion of inducible Cas9 into AAVS1 site of Elf1 cells
10× 106 cells of Elf-1p17 were transfected with 5μg AAVS1-TALEN R plasmid (Addgene #59026), 5μg AAVS1-TALEN L plasmid (Addgene #59025), 40μg Puro-Cas9 donor plasmid (Addgene #58409), 40μg Neo-M2rtTA donor plasmid (Addgene #60843) with Amaxa Lonza Human stem cell Kit #2. The cells were then plated with 5μM of Rock inhibitor (ROCKi) onto 11, 10cm plates onto an irradiated Drug Resistance 4 Mouse embryonic fibroblast feeder cells (DR4). Three days following the nucleofection, the cells were selected for neomycin resistance with Genetecin (50μg/ml) for four days, then selected with Puromycin (0.5μg/ml) for three days. 14 clones survived after the double selection and were isolated and expanded. Of these 14 clones, eight (CL#1,2,4,6,8,11,12,13) clones were plated onto Matrigel with or without doxycycline (2μg/ml) and RNA was extracted in order to analyze the level of Cas9 expression by qPCR. Insertion of iCas9 into the AAVS1 site was confirmed by Southern blot analysis in clones #2, 11 and 12. Clone #2 was sent for G-banded karyotype analysis (Diagnostic Cytogenetics, Seattle) and displayed a normal karyotype.
DNA extraction and sequencing
Genomic DNA was collected using DNAzol reagent (Invitrogen) according to manufacturer's instructions and quantified using Nanodrop ND-1000. Genomic regions flanking the CRISPR target sites were PCR amplified with the designed primers (Table S1), purified by PCR Purification Kit (Invitrogen) and sent to Genewiz for sequencing. Alternatively, following GoTaq PCR amplification of the targeted region, the JARID2 mutations were cloned using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific) following the manufacturer's guidelines.
Southern blot analysis
Southern blot was performed on DNA extracted from iCas9 lines #2, #11 and #12 using probes to detect heterozygous knock-in of inducible CRISPR/Cas9 cassettes into the AAVS1 locus as described previously [Citation5]. Detailed methodology for probe preparation, hybridization and analysis was described previously [Citation22]. The 3′ external and the 5′ internal probes were generated by PCR on plasmid templates using the PCR DIG Probe Synthesis Kit (Roche) using primers 3′F: ACAGGTACCATGTGGGGTTC and 3′R: CTTGCCTCACCTGGCGATAT; 5′F: AGGTTCCGTCTTCCTCCACT and 5′R: GTCCAGGCAAAGAAAGCAAG, respectively. For the 5′ internal probe, we used the Puro-Cas9 donor as a template. BglII digestion of genomic DNA and 3′ external probe hybridization is predicted to yield bands of 12406, 4984 and 7409 bp for wild-type, Puro-Cas9 (or Puro-Cr) and Neo-M2rtTA targeted alleles respectively; whereas SphI digestion and 5′ internal probe hybridization is predicted to yield bands of 6,492, 3,781 and 3,492 bp respectively.
RNA extraction and RT-qPCR analysis
RNA was extracted using Trizol (Life Technologies) according to manufacturer's instructions. RNA samples were treated with Turbo DNase (ThermoFischer) and quantified using Nanodrop ND-1000. Reverse transcription was performed using Random Hexamers (Invitrogen) and Omniscript reverse transcription kit (Qiagen). 10 ng of cDNA were used to perform qRT-PCR using SYBR Green, with suitable primers (Table S2) on an Applied Biosystems 7300 real time PCR system with PCR conditions as stage 1 50°C for 2mins, stage 2 as 95°C for 10 mins, 95°C for 15sec, 60°C for 1 min (40 Cycles). ß-actin was used as an endogenous control.
Statistical analysis
All data are presented as the mean of at least three independent experiments with the standard error of the mean (SEM), unless otherwise indicated. Statistical significance was determined using Student's T-test.
Guide RNA design, synthesis and transfection
The gRNAs targeting the TCTN2, Mel18L, NNMT, HIF1α, HIF2α, PKLR, GPI, IDO1 or JARID2 genes were designed using the CHOPCHOP or CRISPRscan web tools [Citation23,Citation24] (Tables S3) and ordered as T7-gRNA primers. A dsDNA fragment was synthesized from these primers by self annealing PCR to a complementary scaffold primer, which is used to attach the guide to Cas9. The dsDNA fragment was followed by Q5 High Fidelity-based PCR (New England Biolabs). This 120 bp strand served as template for in vitro transcription IVT (MAXIscript T7 kit, applied Biosystems). The RNA was then purified using Pellet Paint® Co-Precipitant (Novagen). The PSEN2 guide was selected using the Zhang Laboratory Target Finder (http://crispr.mit.edu) and the Alt-R CRISPR crRNA guide (5′-CGAUGACGCUGAUCAUGAUG-3′) and tracrRNA were purchased from IDT and the RNA-hybrid was generated using their Alt-R™ CRISPR-Cas9 System. The 147 bp Ultramer DNA repair template was also purchased from IDT (5′-CTCAGCATCTACACGCCATTCACTGAGGACACACCCTCGGTGGGCCAGCGCCTCCTCAACTCCGTGCTGATCACCTTAATCATGATCAGCGTCATCGTGGTTATGACCATCTTCTTGGTGGTGCTCTACAAGTACCGCTGCTACAAG-3′, the mutations introduced into the PSEN2 sequence are underlined). Elf1-iCas9 cells were treated with doxycycline (2μg/ml) for 2 to 3 days before and during transfection. For transfection, cells were dissociated with trypsin, transfected in suspension with gRNAs using Lipofectamine RNAiMAX (Life Technologies) and re-plated onto matrigel-coated plates for NHEJ generated knockouts or onto irradiated MEF plates to generate PSEN2 mutations. gRNA was added at a 40 nM final concentration when added alone and at 15nM when added with 15nM of Ultramer repair template DNA. A second transfection was performed after 24 h. Two days after the last gRNA transfection, Elf1-iCas9 cells were dissociated onto single cells and replated onto MEF-coated plates or collected for DNA analysis. In the next passage single colonies from MEF-coated plates were randomly selected and seeded into a MEF-coated 96 well plate. From the 96 well plate the clones were passaged onto a 24 well on MEF, and finally to 24 well plates on matrigel. The samples were then harvested for protein and analyzed on a Western blot ().
Protein extraction and Western blot analysis
Cells were lysed directly on the plate with lysis buffer containing 20mM Tris-HCl pH 7.5, 150mM NaCl, 15% Glycerol, 1% Triton X-100, 1M ß-Glycerolphosphate, 0.5M NaF, 0.1M Sodium Pyrophosphate, Orthovanadate, PMSF and 2% SDS. 25 U of Benzonase® Nuclease (EMD Chemicals, Gibbstown, NJ) was added to the lysis buffer right before use. Proteins were quantified by Bradford assay (Bio-rad), using BSA (Bovine Serum Albumin) as Standard using the EnWallac Vision. The protein samples were combined with the 4x Laemli sample buffer with 10% β-Mercaptoethanol (Bio-Rad #1610747), heated (95°C, 5mins) and run on SDS-PAGE (protean TGX pre-casted gradient gel, 4%-20%, Bio-rad) and transferred to the Nitro-Cellulose membrane (Bio-Rad) by semi-dry transfer (Bio-Rad). Membrane were blocked for 1hr with 5% milk, and incubated in the primary antibodies overnight in 4°C. The antibodies used for western blot were β-Tubulin III (Promega G7121, 1:1000), JARID2 (Cell Signaling D6M9X, 1:1000), Oct-4 (Santa Cruz sc-5279, 1:1000, Novus Biologicals NB110-90606, 1:500), Nanog (Karol Bomsztyk, University of Washington, 1:1000), H3K27me3 (Active Motive 39155 1:1000), EZH2 (Cell Signaling D2C9, 1:1000) and HIF2α (Abcam, 1:1000). The membranes were then incubated with secondary antibodies (1:10000, goat anti-rabbit or goat anti-mouse IgG HRP conjugate(Bio-Rad) for 1hr and detection was performed using the immobilon-luminol reagent assay (EMP Millipore).
Immunostaining and confocal imaging
Cells were fixed in 4% paraformaldehyde in PBS for 15 min, permeabilized for 10 min in 0.1% Triton X-100 and blocked for 1h in 2% BSA. The cells were then incubated in primary antibody overnight, washed with PBS (3× 5min), incubated with the secondary antibody in 2% BSA for 1hr, washed (4× 10 mins, adding 1μg/ml DAPI in the 2nd wash), mounted (2% of n-Propyl Gallate in 90% Glycerol and 10% PBS) and stored at 4°C. Analysis was done on a Leica TCS-SPE Confocal microscope using a 40x objective and Leica Software. The antibodies for immunostaining were anti-JARID2 (Cell Signaling, 1:200 for 2iL-I-F, 1:100 for 5iLA), anti-Oct-4 (Novus Biologicals, 1:150), anti-Nanog (R&D, 1:200); and Alexa 488- or Alexa 647-conjugated secondary antibodies (Molecular Probes).
Flow cytometry analysis
Cells were dissociated into single cells with Trypsin (Gibco) and fixed using 4% PFA 10 mins at room temperature. Cells were resuspended in PBS with 5%FBS (+0.75% saponin for analysis of intracellular proteins) and incubated with primary antibodies at room temperature for 1h: Oct4 (1:200, Abcam ab19857) or Nanog (1:100, R&DSystems) and washed twice with PBS. Cells were then incubated with isotype-specific secondary antibody (Ig-Alexa 488; Invitrogen) for 30 min. Cells that were analyzed for Tra 1–60 Ab were dissociated into single cells with Trypsin and incubated with Tra 1–60 1:100 (Millipore) in PBS, 5%FBS for 1h on ice, washed twice in PBS and incubated with isotype-specific secondary antibody GaM-PE, 647 for 30min on ice. Unstained cells were used as a control. After staining, cells were resuspended in PBS/5%FBS before analyzing on a BD FACS Canto II. Data analysis was performed with FlowJo Software X 10.0.6 (Tree Star. Inc., Ashland, OR).
Flow cytometry and PSEN2 DNA analysis
A week after transfection cells were dissociated with Trypsin-EDTA, resuspended in PBS with 1% FBS, incubated with TRA-1-81-APC or a mouse IgM-APC isotype control (1:200, 20 min on ice, Affymetrix), centrifuged, and resuspended in HEPES buffered culture media and analyzed on a BD Aria III FACS. TRA-1-81 positive cells were seeded onto 36 wells of a 96 well plate (15 cells/well). A week later 90% of the cells were passaged onto one well of a 12 well plate and the remaining 10% was extracted using QuickExtract DNA Extraction Solution from Epicentre Technologies according to the manufacture's instructions. Two primers (5′- AATGAGCTGGAGGACAGGAACT-3′ & 5′-CAAGACCTCAGCATGGCCTCT-3′) were used to PCR-amplify a 1020 bp fragment containing exon 5 of the PSEN2 gene. The amplified DNA was cut with the restriction enzyme MseI and the digestion products were analyzed on a 1.5% agarose gel. MseI digestion of control DNA is expected to yield restriction fragments of 672,144,133, and 71 bp while DNA repaired with the 147 bp repair template DNA is expected to contain the PSEN2 N141 mutation and an additional MseI restriction site, yielding restriction fragments of 425, 247, 144, 133, and 71 bp. The 3 most positive wells from this analysis (3, 9, and 35) were combined together and single cells were FACS-sorted using TRA-1-81-APC into single wells of a 96 well plate. Eleven clones were recovered on this plate and the clones were passaged and analyzed as described above. All PCR products were sequenced using the forward PCR primer (5′- AATGAGCTGGAGGACAGGAACT-3) by Eurofins MWG Operon.
RNA-seq data analysis
RNA-seq samples from previously published studies were used [Citation11,Citation14,Citation25–27]. RNA-seq samples were aligned to hg19 using Tophat [Citation28] (version 2.0.13). Gene-level read counts were quantified using htseq-count [Citation29] using Ensembl GRCh37 gene annotations. Processed single cell RNA-seq data from Nakamura et al [Citation30] were used. Only genes expressed above 10 Reads Per Million in 3 or more samples were kept. t-SNE [Citation31] was performed with the Rtsne package, using genes with the top 20% variance across samples. Cluster labels from Nakamura et al were used. A Principle Component Analysis (PCA) was performed using all of the cynomolgus monkey samples from Nakamura et al [Citation30] using R software. Genes used in the analysis were restricted to defined homologs expressed at non-zero Transcripts Per Million (TPM) in human in vitro cell lines, and in the preprocessed mouse and cynomolgus monkey single cell samples from Nakamura et al. RNA-seq data from human cell lines were corrected for batch effects using ComBat [Citation32]. Human bulk RNA-seq samples were projected onto the PCA coordinate via matrix multiplication. Human, cynomolgus monkey and mouse RNA-seq data were separately centered and scaled within each species before PCA and projection was performed.
Results
Generation of a stable genetically modified naïve hESC line expressing inducible Cas9
We compared the human pluripotent cell lines and the in vivo non-human primate, cynomolgus monkey (Macaca fascicularis) pre- and post-implantation stage blastocysts to identify naive hESC lines whose gene expression patterns are similar to pre-implantation epiblasts in non-human primate (). We chose one of the Seattle-derived naïve hESC lines (, Suppl. Figure 1A-B,E) to generate an inducible CRISPR/Cas9 stable system in a cell line resembling pre-implantation epiblast. TALEN mediated recombination was then used to insert a doxycycline-inducible Cas9 nuclease expression construct into the human AAVS1 locus in naïve hESC, Elf1. The AAVS1 “safe harbor” locus at 19q13.3 was chosen as the insertion target site since it has an open chromatin configuration that facilitates strong, stable transgene expression in both undifferentiated and differentiated cell states [Citation33–36]. We co-electroporated naïve hESC Elf1 with four plasmids: one containing the inducible-Cas9 cassette and puromycin resistance, one containing a constitutive reverse tetracycline transactivator (M2rtTA) expression cassette and neomycin resistance, and two containing TALEN pairs specific for the AAVS1 site (R and L) [Citation5] (). The targeted cells were then selected using puromycin and neomycin. After selection multiple clonal cell lines were generated (). We assessed the level of Cas9 expression after induction with Doxycycline in 6 of the clonal cell lines by RT-qPCR analysis (Suppl.Figure 1C-D). Three lines showing different levels of conditional expression (lines #2, 11, 12) were further analyzed for Cas9 insertion by Southern blot analysis (). All three lines carried insertion of both donor sequences (iCas9 and M2rtTA, ). Line #2 had no additional random integrations, did not present any leakiness of the construct (Cas9 expression) in cells without Doxycycline treatment (Suppl.Figure 1C-D, Fig. 1E) and displayed normal karyotype (Figure 1F). Line #2 expresses key pluripotent stem cell markers Oct4, Nanog, and TRA-1-60 at similar levels as the original Elf1 line, and the new Seattle-derived naive lines Elf3 and Elf4 (, Suppl.Figure 1A-B, 1E) and was able to exit the pluripotent state when induced to differentiate, similar to the original Elf1 line (Suppl. Figure 1F). In addition, Elf1-iCas9 line #2 can be cultured in various pre-implantation culture conditions (2iL-I-F, 5iLA, EPS [Citation12,Citation18,Citation21]; while retaining expression of stem cell markers (). This newly generated naïve hESC-iCas9 #2 line was therefore chosen for further analysis.
Figure 1. Generation Elf1-iCas9 line. (A) Principle Component Analysis of single-cell RNA-seq data from in vivo samples of cynomolgus monkeys, with human ex vivo blastocyst and in vitro cell lines shown projected onto the principal coordinates ([Citation25-27,Citation30,Citation37]. Only monkey cells of the epiblast lineage (based on cluster labels from [Citation30]) are shown. (B) Schematic representation of the strategy used to insert the inducible Cas9 construct into the AAVS1 locus using a TALEN system in human naive embryonic stem cells Elf1 (adapted from [Citation5]). Blue and orange lines represent the homology of the constructs to PPP1R12C intron 1 on chromosome 19. The black arrows show the cleaving sites of the TALENS; SA, splice acceptor; 2A, self-cleaving 2A peptide; Puro, Puromycin resistance gene; Neo, Neomycin resistance gene; TRE, Tetracyclin responsive element. (C) Workflow of the generation of Elf1-iCas9 lines. (D) Southern blot analysis of lines #2, #11 and #12 using 3′ external and 5′ internal probes. Blue arrow indicates additional random integrations. (E) Elf1-iCas9 cells express Cas9 protein only after doxycycline treatment (2µg/ml for 2 days, Western Blot). (F) Elf1-iCas9 Clone #2 displays a normal G-banded karyotype. (G) Confocal analysis of pluripotent stem cell markers Oct4 and Nanog in the Elf1-iCas9 Clone #2 grown in EPS, 5iLA, and 2iL-I-F conditions. Scale bars represents 50 µm. (H) Flow cytometry analysis of stem cell markers Oct4 and Nanog in Elf1 2iL-I-F and Elf1-iCas9 2i-I-F. The pink peak indicates unstained cells and orange and blue peak represent stained cells for Elf1 and Elf1-iCas9 respectively.
![Figure 1. Generation Elf1-iCas9 line. (A) Principle Component Analysis of single-cell RNA-seq data from in vivo samples of cynomolgus monkeys, with human ex vivo blastocyst and in vitro cell lines shown projected onto the principal coordinates ([Citation25-27,Citation30,Citation37]. Only monkey cells of the epiblast lineage (based on cluster labels from [Citation30]) are shown. (B) Schematic representation of the strategy used to insert the inducible Cas9 construct into the AAVS1 locus using a TALEN system in human naive embryonic stem cells Elf1 (adapted from [Citation5]). Blue and orange lines represent the homology of the constructs to PPP1R12C intron 1 on chromosome 19. The black arrows show the cleaving sites of the TALENS; SA, splice acceptor; 2A, self-cleaving 2A peptide; Puro, Puromycin resistance gene; Neo, Neomycin resistance gene; TRE, Tetracyclin responsive element. (C) Workflow of the generation of Elf1-iCas9 lines. (D) Southern blot analysis of lines #2, #11 and #12 using 3′ external and 5′ internal probes. Blue arrow indicates additional random integrations. (E) Elf1-iCas9 cells express Cas9 protein only after doxycycline treatment (2µg/ml for 2 days, Western Blot). (F) Elf1-iCas9 Clone #2 displays a normal G-banded karyotype. (G) Confocal analysis of pluripotent stem cell markers Oct4 and Nanog in the Elf1-iCas9 Clone #2 grown in EPS, 5iLA, and 2iL-I-F conditions. Scale bars represents 50 µm. (H) Flow cytometry analysis of stem cell markers Oct4 and Nanog in Elf1 2iL-I-F and Elf1-iCas9 2i-I-F. The pink peak indicates unstained cells and orange and blue peak represent stained cells for Elf1 and Elf1-iCas9 respectively.](/cms/asset/bc091f3b-30b1-434b-9dbb-662ee666f312/kccy_a_1442621_f0001_oc.jpg)
Efficient PSEN2 targeting in naïve Elf1-iCas9 line
Homology-directed repair in the Elf1-iCas9 line was used to create the most common disease associated mutation in the PSEN2 gene to study an inherited form of Alzheimer's Disease (AD). The structure and location of AD mutations within the PSEN2 gene are outlined in [Citation38]. A 147 bp DNA repair template was chosen to target the N141 site in exon 5 of PSEN2. This guide added a restriction enzyme cutting site for easy recognition of successful homology-directed repair insertion of the desired mutation (). Cas9 expression was induced in Elf1-iCas9 cells with doxycycline 2 days prior to transfection. Cells were co-transfected with the guide and DNA repair template, and a week later the cells were labeled with TRA-1-81-APC and FAC sorted into 36 wells of a 96 well plate (15 cells/well). After a week, the cells were again passaged and an aliquot of the pool was used to extract genomic DNA, PCR-amplifcation of a 1020 bp genomic fragment of the PSEN2 gene containing exon 5, and digestion of the amplified genomic DNA with the restriction enzyme MseI to assess the degree of editing in the 36 pools of cells (as judged by the appearance of the expected 425 & 247 bp cleavage products). The MseI site in the DNA repair template is only 3 bp downstream of the N141I mutation and so should be tightly linked to the AD mutation (). Analysis of DNA isolated from edited Elf1-iCas9 pools of cells are shown in . Plus signs were used to highlight the relative abundance of the MseI RFLP in pools 1–11, pools 14–24, and pools 25–36. The MseI digest of the DNA derived from these pools of cells was not complete as indicated by the relative abundance of the full-length 1020 bp fragment and the presence of the expected partial digestion product of 348 bp (144+71+133), 215 bp (144+71), and 204 bp (71+133). The pools showing greatest abundance of the MseI RFLP were pooled (wells 3, 9, and 35), stained for TRA-1-81 and FAC sorted a second time into 96 wells of a 96 well plate (1 cell/well) ().
Figure 2. Efficient homology directed repair (HDR) in the Elf1-iCas9 line. (A) Schematic representations of the PSEN2 locus and the relative positions of the disease associated mutations. Reprinted from [Citation22] with permission. (B) Relative location of designed CRISPR guide (arrow) on target exon 5. Repair oligo represented by the block above the guide. Guide and PAM sequence highlighted in gray and mutations in the DNA repair template are shown in lower case text. The MseI site in the DNA repair template is highlighted with a box. (C) Analysis of DNA isolated from edited iCas9 cells after first sort. Cells were sorted into 36 wells of a 96 well plate (15 cells/well). The expected MseI digestion products for control and targeted DNA are: 672,144,133, 71 and 425, 247, 144, 133, 71 bp respectively (arrows). iCas9 cells were induced with doxycycline, transfected with guide RNA plus DNA repair template, stained for TRA-1-81, and then FAC sorted into a 96 well plate (15 cells/well). DNA was isolated and analyzed for the presence of the expected MseI RFLP (a shift of the 672 fragment to 425 + 247 bp). Plus signs were used to highlight the relative abundance of this RFLP in pools 1–11 (), pools 14–24 (), and pools 25–36 (). The MseI digest of the DNA derived from these pools of cells was not complete as indicated by the relative abundance of the full-length 1020 bp fragment and the presence of the expected partial digestion product of 348 bp (144+71+133), 215 bp (144+71), and 204 bp (71+133). (D) Analysis of DNA isolated from clones after the second selection. Edited cells were subjected to two rounds of TRA-1-81-APC FACS. In the second round three pools from the first round (pools 3, 9, 35) were combined and TRA-1-81 FAC sorted a second time into another 96 well plate (1 cell/well). The 11 clones recovered in this second sort are shown, beside two of the prior pools (pool 32 and 35) that serve as a negative and positive control. (E) PSEN2 DNA sequence summary. Exon 5 of the PSEN2 gene was sequenced for each of the 11 targeted clones. Nine of the clones yielded one of the 4 expected genetic compositions, which are summarized above. The N141I mutation is in bold text and the MseI restriction site has been highlighted with a box. The two other clones (clones 1 and 11) both appeared to be heterozygous for the MseI RFLP but upon sequence analysis showed clear evidence of indels at the Cas9 cleavage site.
![Figure 2. Efficient homology directed repair (HDR) in the Elf1-iCas9 line. (A) Schematic representations of the PSEN2 locus and the relative positions of the disease associated mutations. Reprinted from [Citation22] with permission. (B) Relative location of designed CRISPR guide (arrow) on target exon 5. Repair oligo represented by the block above the guide. Guide and PAM sequence highlighted in gray and mutations in the DNA repair template are shown in lower case text. The MseI site in the DNA repair template is highlighted with a box. (C) Analysis of DNA isolated from edited iCas9 cells after first sort. Cells were sorted into 36 wells of a 96 well plate (15 cells/well). The expected MseI digestion products for control and targeted DNA are: 672,144,133, 71 and 425, 247, 144, 133, 71 bp respectively (arrows). iCas9 cells were induced with doxycycline, transfected with guide RNA plus DNA repair template, stained for TRA-1-81, and then FAC sorted into a 96 well plate (15 cells/well). DNA was isolated and analyzed for the presence of the expected MseI RFLP (a shift of the 672 fragment to 425 + 247 bp). Plus signs were used to highlight the relative abundance of this RFLP in pools 1–11 (Figure 2D), pools 14–24 (Figure 2E), and pools 25–36 (Figure 2F). The MseI digest of the DNA derived from these pools of cells was not complete as indicated by the relative abundance of the full-length 1020 bp fragment and the presence of the expected partial digestion product of 348 bp (144+71+133), 215 bp (144+71), and 204 bp (71+133). (D) Analysis of DNA isolated from clones after the second selection. Edited cells were subjected to two rounds of TRA-1-81-APC FACS. In the second round three pools from the first round (pools 3, 9, 35) were combined and TRA-1-81 FAC sorted a second time into another 96 well plate (1 cell/well). The 11 clones recovered in this second sort are shown, beside two of the prior pools (pool 32 and 35) that serve as a negative and positive control. (E) PSEN2 DNA sequence summary. Exon 5 of the PSEN2 gene was sequenced for each of the 11 targeted clones. Nine of the clones yielded one of the 4 expected genetic compositions, which are summarized above. The N141I mutation is in bold text and the MseI restriction site has been highlighted with a box. The two other clones (clones 1 and 11) both appeared to be heterozygous for the MseI RFLP but upon sequence analysis showed clear evidence of indels at the Cas9 cleavage site.](/cms/asset/008332e9-04b4-4aef-9e75-c08019132eac/kccy_a_1442621_f0002_oc.jpg)
Eleven clones were isolated and Exon 5 of the PSEN2 gene was sequenced for each of the 11 targeted clones (Suppl. Figure 2). Nine of the clones yielded one of the 4 expected genetic compositions which are summarized in . Two of the clones were homozygous for both the N141I mutation and the MseI polymorphism (clones 2 & 10), two were heterozygous for both the mutation and the MseI polymorphism (clones 4 and 9), and two lacked both the mutation and the polymorphism (clones 7 and 8). Three clones were heterozygous for the mutation but homozygous for the MseI RFLP (clones 3, 5, and 6). The remaining two clones (clones 1 and 11) both appeared to be heterozygous for the MseI RFLP but upon sequence analysis showed clear evidence of indels at the Cas9 cleavage site.
Efficient NHEJ Indel generation in a naïve Elf1-iCas9 line
To validate Elf1-iCas9 using NHEJ for null mutation, we designed gRNA guides targeting TCTN2, PCGF6(Mel18L), NNMT, HIF1α, HIF2α, IDO1, PKLR, GPI, and JARID2 genes using the CHOPCHOP and CRISPRscan web tools [Citation23,Citation24]. Following transfections of Elf1-iCas9 with gRNA in presence of Doxycycline, DNA was isolated from cell pool, PCR amplified around the editing site and subjected to Sanger sequencing to test the mutation efficiency of the guide in the pool (). If the sequence trace of the pool revealed a mixture of wild type sequence and InDel mutations starting at the Cas9 cleavage site, 3 nucleotides upstream of the PAM sequence, the guide was considered efficient in generating Cas9 based mutations. All guides shown induced indels in the hESC pools for the chosen genes in Elf1-iCas9 (clone #2) line, however, the frequency of indels varied depending on the guide (). If no sequence alterations were observed around PAM sites, the guide was considered non-functional. Guides scoring well with the CRISPRscan web tool induced InDel mutations efficiently (, ) while guides scoring poorly with this tool were less efficient ().
Figure 3. Efficient NHEJ Indel generation in naïve Elf1-iCas9 line. (A) Schematic representation of knock out generation by InDel mutations. (B-H) Relative location of designed CRISPR guides on schematic representation of TCTN2 (B), Mel18L (C), NNMT (D), HIF1α (E), IDO1 (F), PKLR (G), GPI (H) proteins and Sanger sequencing trace file of the pools of Elf1-iCas9 cells transfected with sgRNA. The area covered by the guide is shown in dark blue. Both mutant and wild-type sequences are shown. Black arrow indicates start of mutated region
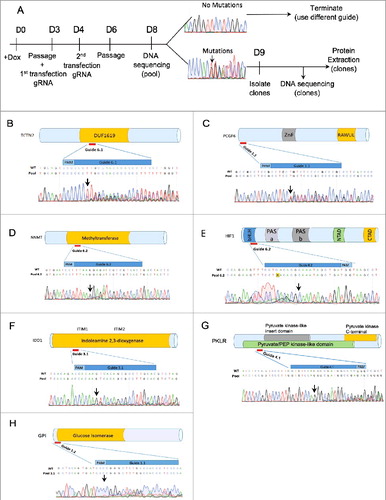
Functional guides efficiently induce mutation in Elf1-iCas9 cells when cultured in various naïve conditions (2iL-I-F, 5iLA [Citation12,Citation21] (), demonstrating the utility of this platform to study early stages of human pluripotency.
Figure 4. Efficiency of gene targeting in Elf1-iCas9 line. (A) Timeline of the experimental procedure. (B) Schematic representation of the JARID2 protein and relative localization of guides 3.1, 4.4 and 6.1 on DNA sequence. JmjN: Jumonji domain N; ARID: AT-rich interaction domain; ZnF: Zinc Finger domain; K116: EED binding residue. Sanger sequencing trace file of the pool of Elf1-iCas9 cells grown in either 2iL-I-F or 5iLA naïve media and transfected with JARID2 sgRNA 6.1, or 3.1. The area covered by the guide is shown in dark blue. Both mutant and wild-type sequences are shown. Black arrows indicate start of the mutated region. (C) Western Blot analysis for JARID2 Knockouts and Knockdowns in Elf1-iCas9 cells. (D) Western Blot analysis for HIF2α Knockout and Knockdown in Elf1-iCas9 cells (left panel) and schematic representation of HIF2α on right panel. (E) Table showing mutagenesis efficiency of JARID2 gRNA 6.1(exp.1), JARID2 gRNA 3.1(exp. 2) and HIF2α gRNA 5 in Elf1-iCas9 line.
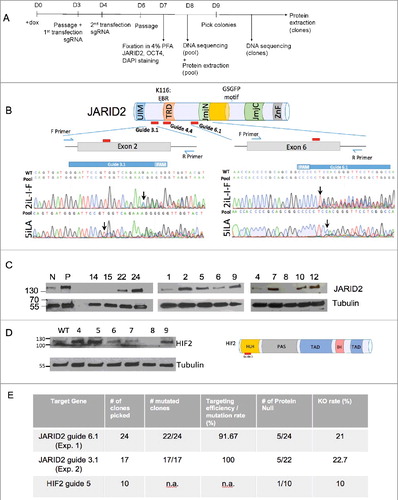
To isolate mutant colonies and to quantify the targeting efficiency and InDel formation, clones were isolated from the pool of mutant cells for JARID2 (, Suppl. Figure 3). The individual clones were analyzed for mutations by Sanger sequencing (Suppl.Figure 3). Out of 48 isolated JARID2 clones, 46 displayed DNA mutations in the guide region. Analysis of the top predicted off-target sites for the JARID2 guide did not detect any mutation, neither in the pool nor individual clones (Suppl. Figure 4). The high InDel rate (96%) observed in the guide region using JARID2 guides further demonstrated a high CRISPR targeting efficiency in the Elf1-iCas9 #2 line (Figure 4E). However, while mutation rate using this iCas9 line with one guide system is very high, only 10 out of 46 JARID2 mutant clonal lines (22%) resulted in protein-null mutations (Figure 4C, 4E). Similarly, we also generated HIF2α protein null mutants and evaluated efficiency in Elf1-iCas9 line (Figure 4D). After transfections with guides targeting HIF2α, we generated clonal lines. To identify the clones with mutations in HIF2α protein, we exposed the cells to 2% O2 for 4 hours to stabilize HIF2α protein. One out of ten clones analyzed showed no HIF2α protein expression, suggesting that the protein null clonal frequency for HIF2α was 10% using Elf1-iCas9 (#2) line ().
JARID2 expression in pluripotent stages
JARID2 is one of the members of the Polycomb group proteins and plays a role in generating the repressive histone methylation, H3K27me3 marks by PRC2 [Citation39–43]. The JARID2 protein contains an Ubiquitin Interactive Motif (UIM), transcription repressive domain (TRD), inactive histone demethylase domains (JmjN, JmjC), DNA binding domains (ARID, ZnF) and the ‘GSGFP′ Suz12 binding motif () [Citation43–45]. Mouse naïve ECS express pluripotency markers even if components of PRC2 have been eliminated [Citation46,Citation47], suggesting that repressive H3K27me3 marks are dispensable for the earliest stabilized pluripotent stage. Since multiple pluripotent stages have been stabilized from early human development [Citation10–14], it is critical to analyze if PRC2 components are required for stemness in these stages. Using a computationally designed new protein, EED binder, we recently showed that similar to mouse, the earliest naïve 5iLA hESC state is PRC2-independent, however, PRC2 is required to maintain naïve 2iL-I-F and primed hESC pluripotent states [Citation20]. Therefore, PRC2 is not required in earliest naïve state in either mouse or in human, but is required in later stages [Citation20].
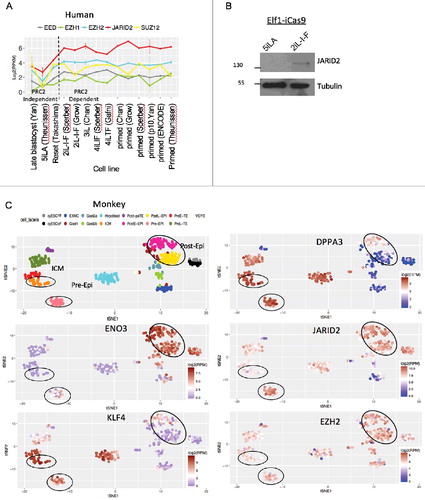
To address the mechanism for the PRC2 differential requirement between naïve 5iLA and later pluripotent states, we analyzed the expression of PRC2 components in different pluripotent stages in cell culture and in vivo, utilizing the stabilized human ECS lines, and pre- and post-implantation in vivo blastocysts from non-human primate, cynomolgus monkey (Macaca fascicularis). Among PRC2 components, JARID2 expression showed the most significant change when the pluripotent cells exit the PRC2 independent, 5iLA naive state and enter the PRC2 dependent, 2iL-I-F state (). To examine the expression heterogeneity of PRC2 components in pre- and post-implantation in vivo blastocysts, we performed t-distributed stochastic neighbor embedding analysis (t-SNE) of in vivo monkey single cell RNA-seq data [Citation30]. t-SNE can capture the non-linear trend and is more robust to noise in data. It is a commonly used clustering method in single cell data analysis. t-SNE captured distinct cell types along pre- and post-implantation development. We first validated known naïve ESC markers [Citation48] and ENO3, a target of primed marker HIF1α [Citation12] using t-SNE. We then analyzed PRC2 component expression in these cell types (). JARID2 is observed in ICM, pre-Epi and post-Epi stages. However, JARID2 expression is dramatically increased between ICM and pre-implantation epiblast stage in non-human primate development (). Expression of other components of PRC2 complex is either stable or slightly up-regulated between pre-implantation and post-implantation stages (). These data show that in both stabilized pluripotent cell cultures and in in vivo blastocysts JARID2 expression levels are increased after the earliest pluripotent stages. Since previous studies show that JARID2's tri-methylated lysine-116 binds to the aromatic cage of the EED, suggesting that JARID2 functions as a potential recruiter for PRC2 based repression of transcription [Citation49], dramatic change in JARID2 expression levels during pluripotency stages may be causal for changes in PRC2 function. We therefore tested if JARID2 is required for human pluripotency.
Figure 5. PRC2 components in human and non-human primate pluripotent stem cells in vitro and in vivo. (A) Expression of PRC2 components across human ex vivo and in vitro ESC at different stages of pluripotency. Error bars represent the standard deviation. (B) Western Blot analysis of JARID2 protein in Elf1-iCas9 culture in 5iLA and 2iL-I-F conditions. (C) t-SNE clusters of monkey single cell RNA-seq data and expression of pre- and post-implantation markers, and PRC2 components. Each dot represents a single cell.
Characterization of JARID2 KO phenotype in naïve hESC
To test for JARID2 function in naïve 2iL-I-F hESC, we analyzed the clones with null expression of JARID2 protein for H3K27me3 marks and EZH2 expression by Western blotting. The clonal lines exhibited early stop codons in the JARID2 gene, but showed no obvious off-target mutations (; Suppl.Figure 4B and 5). Dramatic downregulation of H3K27me3 and EZH2 was observed in all JARID2 knockout clones (). The downregulation of EZH2 might be the result of degradation since it has previously been shown that disruption of the PRC2 complex leads to the degradation of the EZH2 protein [Citation20,Citation50].
Figure 6. JARID2 KO phenotype in naïve hESC. (A) Western blot analysis of EZH2, OCT4, NANOG, and H3K27Me3 marks in JARID2 Knockout clones of 2iL-I-F naïve hESC. (B) Schematic representation of mutation in JARID2 KO Clone2. (C-D) Confocal imaging of wild type Elf1-iCas9 cells and Elf1-iCas9 cells transfected with JARID2 sgRNA6.1 (2iL-I-F(C); 5iLA(D)). Blue: DAPI; Green: Oct4, Red: Nanog and Magenta: JARID2 (higher concentration of Ab used for 5iLA to detect lower levels of JARID2: JARID2 Ab 1:200 in 2iL-I-F; 1:100 in 5iLA). The arrows indicate JARID2 KO cells, Scale bar 50 µm (C) or 10 µm (D).
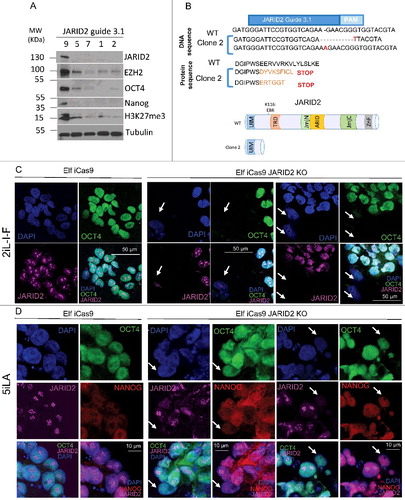
To test if reduction of repressive H3K27me3 marks, due to JARID2 KO, affected pluripotency, we analyzed OCT4 and NANOG protein expression in the JARID2 KO hESC lines with reduced EZH2 and H3K27me3 levels (). All JARID2 hESC KO lines lacked or showed dramatically reduced OCT4 and NANOG expression (), an indication of reduced pluripotency. Accordingly, these lines lost their stem cell colony morphology and the capacity to be propagated in stem cell growth conditions.
To further probe the relationship between JARID2 and OCT4 in different stages of pluripotency of hESC, and to control for potential guide related unspecific background effects, we used a second guide to make mutations in exon6 in the JARID2 gene () and analyzed the cellular effect in OCT4 expression in 2iL-I-F and 5iLA conditions. We performed immunofluorescence for JARID2 and OCT4 proteins immediately after induction of mutations in Elf1-iCas9 2iL-I-F and 5iLA lines using the JARID2 guide 6.1 (). Confocal analysis of these JARID2 knock-out pools of cells revealed that while wild type clones express JARID2 and OCT4, the JARID2 knockout cells lacked OCT4 staining in the 2iL-I-F state, but had normal nuclear OCT4 and NANOG staining in the 5iLA state (, arrows). These data suggest that 2iL-I-F, but not 5iLA hESC that lack JARID2 have highly reduced expression of the pluripotency marker OCT4. However, more careful analysis is required in order to dissect in detail the role of JARID2 in various stages of human pluripotency.
Discussion
Here we report the generation of a naïve hESC line that contains an inducible Cas9 construct in the AAVS1-site (Elf1-iCas9). Mutations in this line were efficiently generated in nine genes. Doxycycline induction of Cas9 can be used for CRISPR-based mutant screens and further, to generate human disease models in hESC. This approach should be broadly applicable to naïve hESC differentiation paradigms.
Using the Elf1-iCas9 line we generated heterozygous and homozygous PSEN2N141I missense mutations. Heterozygous mutations in PSEN2 cause familial Alzheimer's Disease (AD) with nearly 100% penetrance [Citation38], though the mechanisms by which the PSEN2N141l mutation causes neurodegeneration in patients is not known. Murine models have been valuable for the study of AD pathogenesis, however, genomic differences between mouse and human remains a barrier to translating cell biology insights gleaned in animal models. Thus there is significant interest in developing neural tissue models to recapitulate disease in humans [Citation51]. Here we have developed an AD hESC line that can be further differentiated into AD relevant cell types and exploited to investigate cell autonomous and non-cell autonomous mechanisms. Furthermore, our platform allows for assessment of relative impact of various disease-associated mutations within the same genomic background thus circumventing confounders of genomic heterogeneity between individuals.
We next utilized the new Elf1-iCas9 line to study the regulation of PRC2 dependent epigenetic modifications in hESC [Citation20], by generating mutations in the PRC2 component JARID2. Two major polycomb complexes in mammals, polycomb repressive complex 1 (PRC1) and 2 (PRC2), interact with chromatin to sequentially establish distinct post-translational modifications of histone tails, which determine the degree of transcriptional repression at specific loci. The mammalian PRC2 complex is established by the interactions of the EZH2 (or EZH1), EED, Suz12, and RBAP46/48 subunits [Citation52–58]. Genome-wide approaches revealed that H3K27me3 is widely distributed among genes encoding developmental regulators [Citation59–64]. JARID2 (jumonji, AT rich interactive domain 2) is a nuclear protein necessary for mouse embryogenesis [Citation41,Citation44,Citation45,Citation57,Citation65]). JARID2 contains a DNA-binding domain (the AT-rich interaction domain, ARID), a zinc finger domain, a jumonji N (JmjN) domain and a JmjC domain [Citation43–45]. Many JmjC domain-containing proteins have been shown to catalyze lysine demethylation, however, the residues required for iron and ascorbate binding, essential for demethylation activity, are not conserved in the JmjC domain of JARID2 [Citation66]. JARID2 can interact with PRC2, stimulating H3K27 methylation activity, has a DNA-binding activity that is not dependent on the ARID domain only, and facilitates PRC2 recruitment to its target genes [Citation1,Citation41,Citation44,Citation45,Citation49]. Moreover, JARID2 is required for PRC2-repressive activity in mouse ES cells, as evidenced by their impaired differentiation in its absence [Citation45]. JARID2's tri-methylated lysine-116 binds to the aromatic cage of the EED (Embryonic Ectoderm Development), suggesting that JARID2 functions as a potential recruiter for PRC2 based repression of transcription [Citation49].
The PRC2 dependent H3K27me3 repressive marks are significantly upregulated during the implantation stage of early embryonic development [Citation10,Citation12,Citation13,Citation20]. However, it was not known if JARID2 is required for the early H3K27me3 marks in hESC. We have now analyzed the relationship between JARID2 and PRC2-dependent histone methylation (H3K27me3) marks and shown that JARID2 KO in 2iL-I-F hESC reduces EZH2 protein levels and H3K27me3 marks, revealing the function of JARID2 in hESC PRC2 at the 2iL-I-F pluripotency stage. Since JARID2 functions as a potential recruiter for PCR2 [Citation63], the data suggest that JARID2 may play a critical regulatory function at the stage of pre-implantation pluripotency when PRC2 is first required. Future experiments are directed towards revealing the key gene loci for JARID2 localization in 2iL-I-F hESC. These experiments suggest and are warranted to further test if JARID2 is responsible for differential PRC2 dependency during early human pluripotency [Citation20]. Furthermore, since recent findings suggest that JARID2 acts as a linker/regulator between PRC1 and PRC2 [Citation43], it will be informative to analyze the function of PRC1 in naïve hESC.
Disclosure of potential conflicts of interest
The authors declare no conflicts of interest.
Supplemental Material
Download PDF (10.2 MB)Acknowledgments
We thank members of the Ruohola-Baker laboratory for helpful discussions throughout this work. We thank Kristin Goodsell, Varun Ramaswamy, Jason Miklas, and Dannie Macrin for help throughout this study. We thank Dr. Karol Bomsztyk for providing Nanog antibody. This work is supported by grants from the National Institutes of Health R01GM097372, R01GM97372-03S1 and R01GM083867 for HRB, 1P01GM081619 for CW, CM and HRB, T32CA080416 for AF, and the NHLBI Progenitor Cell Biology Consortium (U01HL099997; UO1HL099993) for PP, CW and HRB.
Additional information
Funding
References
- Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19(3):193–204. doi:10.1634/stemcells.19-3-19310.1634/stemcells.19-3-193. PMID: 11359944
- Muffat J, Li Y, Jaenisch R. CNS disease models with human pluripotent stem cells in the CRISPR age. Curr Opin Cell Biol. 2016;43:96–103. doi:10.1016/j.ceb.2016.10.00110.1016/j.ceb.2016.10.001. PMID: 27768957
- Muffat J, Li Y, Yuan B, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22(11):1358–1367. doi:10.1038/nm.418910.1038/nm.4189. PMID: 27668937
- Waddington SN, Privolizzi R, Karda R, et al. A Broad Overview and Review of CRISPR-Cas Technology and Stem Cells. Curr Stem Cell Reports. 2016;2(1):9–20. doi:10.1007/s40778-016-0037-510.1007/s40778-016-0037-5.
- González F, Zhu Z, Shi Z-D, et al. An iCRISPR Platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15(2):215–226.https://doi.org/10.1016/j.stem.2014.05.01810.1016/j.stem.2014.05.018. PMID: 24931489
- Chen Y, Cao J, Xiong M, et al. Engineering human stem cell lines with inducible gene knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17(2):233–244. doi:10.1016/j.stem.2015.06.00110.1016/j.stem.2015.06.001. PMID: 26145478
- Mandegar MA, Huebsch N, Frolov EB, et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell. 2016;18(4):541–553. doi:10.1016/j.stem.2016.01.02210.1016/j.stem.2016.01.022. PMID: 26971820
- Bertero A, Pawlowski M, Ortmann D, et al. Optimized inducible shRNA and CRISPR/Cas9 platforms for in vitro studies of human development using hPSCs. Development. 2016;143(23):4405–4418. doi:10.1242/dev.13808110.1242/dev.138081. PMID: 27899508
- Ware CB, Nelson AM, Mecham B, et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci USA. 2014;111(12):4484–4489. doi:10.1073/pnas.131973811110.1073/pnas.1319738111.
- Theunissen TW, Powell BE, Wang H, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15(4):471–487. doi:10.1016/j.stem.2014.07.00210.1016/j.stem.2014.07.002. PMID: 25090446
- Takashima Y, Guo G, Loos R, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158(6):1254–1269. doi:10.1016/j.cell.2014.08.02910.1016/j.cell.2014.08.029. PMID: 25215486
- Sperber H, Mathieu J, Wang Y, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol. 2015;17(12):1523–1535. doi:10.1038/ncb326410.1038/ncb3264. PMID: 26571212
- Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504(7479):282–286. doi:10.1038/nature1274510.1038/nature12745. PMID: 24172903
- Chan Y-S, Göke J, Ng J-H, et al. Induction of a Human Pluripotent State with Distinct Regulatory Circuitry that Resembles Preimplantation Epiblast. Cell Stem Cell. 2013;13(6):663–675. doi:10.1016/j.stem.2013.11.01510.1016/j.stem.2013.11.015. PMID: 24315441
- Qin H, Hejna M, Liu Y, et al. YAP induces human naive pluripotency. Cell Rep. 2016;14(10):2301–2312. doi:10.1016/j.celrep.2016.02.03610.1016/j.celrep.2016.02.036. PMID: 26947063
- Pastor WA, Chen D, Liu W, et al. Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell. 2016;18(3):323–329. doi:10.1016/j.stem.2016.01.01910.1016/j.stem.2016.01.019. PMID: 26853856
- Guo G, von Meyenn F, Santos F, et al. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports. 2016;6(4):437–446. doi:10.1016/j.stemcr.2016.02.00510.1016/j.stemcr.2016.02.005. PMID: 26947977
- Yang Y, Liu B, Xu J, et al. Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell. 2017;169(2):243–257. doi:10.1016/j.cell.2017.02.00510.1016/j.cell.2017.02.005. PMID: 28388409
- Yang Y, Zhang X, Yi L, et al. Naive induced pluripotent stem cells generated from -thalassemia fibroblasts allow efficient gene correction with CRISPR/Cas9. Stem Cells Transl Med. 2016;5(2):267-267. doi:10.5966/sctm.2015-0157erratum10.5966/sctm.2015-0157erratum. PMID: 26819338
- Moody JD*, Levy S*, Mathieu J*, et al. First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor. PNAS. 2017;114(38):10125–10130. doi:10.1073/pnas.170690711410.1073/pnas.1706907114. PMID: 28864533
- Theunissen TW, Friedli M, He Y, et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19(4):502–515. doi:10.1016/j.stem.2016.06.01110.1016/j.stem.2016.06.011. PMID: 27424783
- Chong JJH, Yang X, Don CW, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510(7504):273–277. doi:10.1038/nature1323310.1038/nature13233. PMID: 24776797
- Montague TG, Cruz JM, Gagnon JA, et al. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42(Web Server issue):W401–W407. doi:10.1093/nar/gku41010.1093/nar/gku410. PMID: 24861617
- Moreno-Mateos MA, Vejnar CE, Beaudoin J-D, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12(10):982–988. doi:10.1038/nmeth.354310.1038/nmeth.3543. PMID: 26322839
- Yan L, Yang M, Guo H, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20(9):1131–1139. doi:10.1038/nsmb.266010.1038/nsmb.2660. PMID: 23934149
- Theunissen TW, Friedli M, He Y, et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19(4):502–515. doi:10.1016/j.stem.2016.06.01110.1016/j.stem.2016.06.011. PMID: 27424783
- Grow EJ, Flynn RA, Chavez SL, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522(7555):221–225. doi:10.1038/nature1430810.1038/nature14308. PMID: 25896322
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi:10.1093/bioinformatics/btp12010.1093/bioinformatics/btp120. PMID: 19289445
- Novodvorska M, Stratford M, Blythe MJ, et al. Metabolic activity in dormant conidia of Aspergillus niger and developmental changes during conidial outgrowth. Fungal Genet Biol. 2016;94:23–31. doi:10.1016/j.fgb.2016.07.00210.1016/j.fgb.2016.07.002. PMID: 27378203
- Nakamura T, Okamoto I, Sasaki K, et al. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537(7618):57–62. doi:10.1038/nature1909610.1038/nature19096. PMID: 27556940
- Van Der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605.
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi:10.1093/biostatistics/kxj03710.1093/biostatistics/kxj037. PMID: 16632515
- Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27(9):851–857. doi:10.1038/nbt.156210.1038/nbt.1562. PMID: 19680244
- Mathieu J, Zhang Z, Nelson A, et al. Hypoxia induces re-entry of committed cells into pluripotency. Stem Cells. 2013;31(9):1737–1748. doi:10.1002/stem.144610.1002/stem.1446. PMID: 23765801
- Gantz JA, Palpant NJ, Welikson RE, et al. Targeted genomic integration of a selectable floxed dual fluorescence reporter in human embryonic stem cells. PLOS One. 2012;7(10):e46971. doi:10.1371/journal.pone.004697110.1371/journal.pone.0046971. PMID: 23071682
- Smith JR, Maguire S, Davis LA, et al. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26(2):496–504. doi:10.1634/stemcells.2007-003910.1634/stemcells.2007-0039. PMID: 18024421
- Takashima Y, Guo G, Loos R, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158(6):1254–1269. doi:10.1016/j.cell.2014.08.02910.1016/j.cell.2014.08.029. PMID: 25215486
- Jayadev S, Leverenz JB, Steinbart E, et al. Alzheimer's disease phenotypes and genotypes associated with mutations in presenilin 2. Brain. 2010;133(4):1143–1154. doi:10.1093/brain/awq03310.1093/brain/awq033. PMID: 20375137
- Vizán P, Beringer M, Ballaré C, et al. Role of PRC2-associated factors in stem cells and disease. FEBS J. 2015;282(9):1723–1735. doi:10.1111/febs.1308310.1111/febs.13083. PMID: 25271128
- Landeira D, Bagci H, Malinowski AR, et al. Jarid2 Coordinates nanog expression and PCP/Wnt signaling required for efficient ESC differentiation and early embryo development. Cell Rep. 2015;12(4):573–586. doi:10.1016/j.celrep.2015.06.06010.1016/j.celrep.2015.06.060. PMID: 26190104
- Pasini D, Cloos PAC, Walfridsson J, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–310. doi:10.1038/nature0878810.1038/nature08788. PMID: 20075857
- Zhang Z, Jones A, Sun C-W, et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprograming. Stem Cells. 2011;29(2):229–240. doi:10.1002/stem.57810.1002/stem.578. PMID: 21732481
- Cooper S, Grijzenhout A, Underwood E, et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun. 2016;7:13661. doi:10.1038/ncomms1366110.1038/ncomms13661. PMID: 27892467
- Peng JC, Valouev A, Swigut T, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi:10.1016/j.cell.2009.12.00210.1016/j.cell.2009.12.002. PMID: 20064375
- Li G, Margueron R, Ku M, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24(4):368–380. doi:10.1101/gad.188641010.1101/gad.1886410. PMID: 20123894
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26(6):1496–1505. doi:10.1634/stemcells.2008-010210.1634/stemcells.2008-0102. PMID: 18403752
- Galonska C, Ziller MJ, Karnik R, et al. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell. 2015;17(4):462–470. doi:10.1016/j.stem.2015.07.00510.1016/j.stem.2015.07.005. PMID: 26235340
- Weinberger L, Ayyash M, Novershtern N, et al. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17(3):155–169. doi:10.1038/nrm.2015.2810.1038/nrm.2015.28. PMID: 26860365
- Sanulli S, Justin N, Teissandier A, et al. Jarid2 Methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol Cell. 2015;57(5):769–783. doi:10.1016/j.molcel.2014.12.02010.1016/j.molcel.2014.12.020. PMID: 25620564
- Collinson, A., et al., Deletion of the Polycomb-Group Protein EZH2 Leads to Compromised Self-Renewal and Differentiation Defects in Human Embryonic Stem Cells. Cell Rep. 2016;17(10):2700–2714. doi:10.1016/j.celrep.2016.11.03210.1016/j.celrep.2016.11.032. PMID: 27926872
- Young JE, Goldstein LSB. Alzheimer's disease in a dish: promises and challenges of human stem cell models. Hum Mol Genet. 2012;21(R1):R82–R89. doi:10.1093/hmg/dds31910.1093/hmg/dds319. PMID: 22865875
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. (80-) 2002;298(5595.): doi:10.1126/science.107699710.1126/science.1076997.
- Czermin B, Melfi R, McCabe D, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111(2):185196. doi:10.1016/S0092-8674(02)00975-310.1016/S0092-8674(02)00975-3. PMID: 12408863
- Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18(13):1592–1605. doi:10.1101/gad.120020410.1101/gad.1200204. PMID: 15231737
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15(1):57–67. doi:10.1016/j.molcel.2004.06.02010.1016/j.molcel.2004.06.020. PMID: 15225548
- Margueron R, Li G, Sarma K, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32(4):503–518. doi:10.1016/j.molcel.2008.11.00410.1016/j.molcel.2008.11.004. PMID: 19026781
- Shen X, Kim W, Fujiwara Y, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi:10.1016/j.cell.2009.12.00310.1016/j.cell.2009.12.003. PMID: 20064376
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, et al. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi:10.1101/gad.103590210.1101/gad.1035902. PMID: 12435631
- Bernstein E, Duncan EM, Masui O, et al. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26(7):2560–2569. doi:10.1128/MCB.26.7.2560-2569.200610.1128/MCB.26.7.2560-2569.2006. PMID: 16537902
- Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi:10.1038/nature0473310.1038/nature04733. PMID: 16625203
- Bracken AP, Dietrich N, Pasini D, et al. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20(9):1123–1136. doi:10.1101/gad.38170610.1101/gad.381706. PMID: 16618801
- Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi:10.1016/j.cell.2006.02.04310.1016/j.cell.2006.02.043. PMID: 16630818
- Schwartz YB, Kahn TG, Nix DA, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38(6):700–705. doi:10.1038/ng181710.1038/ng1817. PMID: 16732288
- Squazzo SL, O'Geen H, Komashko VM, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16(7):890–900. doi:10.1101/gr.530660610.1101/gr.5306606. PMID: 16751344
- Landeira D, Sauer S, Poot R, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12(6):618–624. doi:10.1038/ncb206510.1038/ncb2065. PMID: 20473294
- Takeuchi T, Watanabe Y, Takano-Shimizu T, et al. Roles ofjumonji andjumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235(9):2449–2459. doi:10.1002/dvdy.2085110.1002/dvdy.20851. PMID: 16715513
