ABSTRACT
The general snRNA gene transcription apparatus has been extensively studied. However, the role of coactivators in this process is far from being clearly understood. Here, we have demonstrated that the Drosophila SAGA complex interacts with the PBP complex, the key component of the snRNA gene transcription apparatus, and is present at the promoter regions of the snRNA genes transcribed by both the RNA polymerase II and RNA polymerase III (U6 snRNA). We show that SAGA interacts with the Brf1 transcription factor, which is a part of the RNA polymerase III transcription apparatus and is present at promoters of a number of Pol III-transcribed genes. Mutations inactivating several SAGA subunit genes resulted in reduced snRNA levels in adult flies, indicating that SAGA is indeed the transcriptional coactivator for the snRNA genes. The transcription of the Pol II and Pol III-transcribed U genes was reduced by mutations in all tested SAGA complex subunits. Therefore, the transcription of the Pol II and Pol III-transcribed U genes was reduced by the mutations in the deubiquitinase module, as well as in the acetyltransferase module of the SAGA, indicating that the whole complex is essential for their transcription. Therefore, the SAGA complex activates snRNA genes suggesting its wide involvement in the regulation of gene transcription, and consequently, in the maintenance of cellular homeostasis.
Introduction
The snRNAs are non-coding RNA molecules expressed at high levels in eukaryotic cells. snRNAs are involved in many cellular functions including pre-mRNA splicing, rRNA processing, and histone mRNA 3’-end formation. While for many years the snRNA have been frequently described as mostly performing housekeeping functions, however their real functions extend beyond housekeeping, and their regulated expression is critical to cellular homeostasis during normal growth and in response to stress [Citation1]. Each snRNA gene is represented by several copies which are organised in clusters or scattered throughout the genome. In animals, most snRNAs, namely, U1, U2, U3, U4, U5, and U7, are synthesised by the RNA polymerase II, while the U6 snRNA is synthesised by the RNA polymerase III [Citation1–Citation4]. Although Drosophila snRNA genes are transcribed by different RNA polymerases, they have similarly organised promoters containing the distal sequence element (DSE), the transcription enhancer, and the proximal sequence element (PSE, or more specifically, PSEA in Drosophila), essential for basal transcription [Citation5–Citation7]. Drosophila snRNA promoters directing the transcription by Pol II contain, apart from the PSEA element, the PSEB element which is also required for basal transcription. In promoters transcribed by Pol III, TATA-box is present in addition to PSEA, which determines the specificity of the Pol III recruitment [Citation5].
The PSEs of all snRNA genes are recognised and bound by the same evolutionarily conserved PBP factors, also known as the SNAP factors [Citation8].
The interaction between PSE and PBP induces the recruitment of RNA polymerases to the gene [Citation1,Citation5]. The Drosophila PBP complex contains three subunits, Pbp95 (Snap190), Pbp49 (Snap50) and Ppb45 (Snap43) [Citation5,Citation9]. Apart from the PBP factors, the basal transcription of snRNA genes by the RNA polymerase II engages TBP, TFIIA, TFIIB, TFIIF, and TFIIE. The basal transcription of the RNA polymerase III-transcribed snRNA genes involves in addition to PBP, also TBP, Bdp1, and Brf1 (BRF2 in human) [Citation10,Citation11].
The SAGA complex is currently known as the transcriptional coactivator within the RNA polymerase II transcription machinery [Citation12]. Histone acetylation has for a long time been associated with active gene transcription. This modification is dynamically regulated by the counteracting histone acetyltransferases (HAT) and deacetylases, whose targets are a number of highly conserved residues in the N-terminal amino acid sequences of histones. In Drosophila, similar to other organisms, SAGA consists of several functional modules, including the histone acetylation module (HAT), which contains the Gcn5 acetyltransferase along with the Ada and Sgf29 proteins, and the deubiquitination module (DUB), containing the Nonstop deubiquitinase as well as the Sgf11, Ataxin7 and ENY2 subunits [Citation12–Citation16]. Recently, it was shown that subunits of U2 snRNP: SF3B3 and SF3B5 have independent functions in SAGA [Citation17,Citation18]. The best characterised substrates for the SAGA complex are several acetylation sites in histone H3 and a single deubiquitination site in histone H2B [Citation19,Citation20]. SAGA also contains two structural modules, TAF and SPT, which ensure the structural integrity of the complex and are essential for the interaction with the general transcription machinery. The composition of the SAGA complex, which is also referred to as STAGA or TFTC, is to a large degree conserved across different species [Citation21]. In Drosophila, there has been described an evolutionarily related ATAC complex, which shares a part with SAGA [Citation22,Citation23]. These two complexes share a number of common subunits, such as dTada3, dGcn5, and dSgf29, but differ by other ones. One of the major differences between them is the absence of the deubiquitinase module in the ATAC complex [Citation18]. Several reviews covering the composition and structure of the SAGA complex, as well as the differences between the SAGA-like complexes, have been published recently [Citation18,Citation24]. A number of studies have reported that the Gcn5 and TRRAP subunits of the SAGA complex are also engaged in transcription of the RNA polymerase III-dependent genes such as tRNA and 5S RNA. These factors are recruited to the transcription apparatus via c-Myc and perform functions similar to those performed during the RNA polymerase II transcription [Citation25]. Recently, Li et al, investigating SAGA-independent properties of the DUB module, detected by a ChIP-seq assay an occupancy of snRNA promoter regions by Sgf11, Spt3, and Ada2b subunits [Citation26].
The regulation of snRNA gene transcription is now intensively studied. Recently, it has been demonstrated that Mediator subunits are involved in the snRNA gene transcription controlled by the RNA polymerase II [Citation27,Citation28]. However, the functions of coactivator complexes in snRNA transcription remain poorly understood. The current work reports the study of the role of the SAGA complex in this process.
Results
SAGA is present at snrna genes and interacts with pbp45
Mapping the binding sites for Sgf11 component of the SAGA complex on the polytene chromosomes of Drosophila by immunostaining revealed that Sgf11 is present at the sites of localization of snRNA genes [Citation29]. To verify this result, we produced rabbit polyclonal antibodies against the Pbp45 protein (Supplementary Figure1(a)), the subunit of the PBP complex, the key player in the snRNA transcription process. The antibodies were affinity purified and their specificity was confirmed by RNAi knockdown of Pbp45 (Supplementary Figure 1(c)).
Figure 1. SAGA is colocalised with Pbp45 at many sites on polytene chromosomes of Drosophila. Sgf11 (green) colocalised with Pbp45 (red) on polytene chromosomes of Drosophila in the loci, corresponding to snRNA genes. The Drosophila chromosomes (X, 2L, 2R, 3L and 3R) are indicated. Chromosomes were stained with anti-Sgf11 antibodies (a), with anti-Pbp45 antibodies (b), and co-stained with DAPI (d). Merged image is shown (c). Arrows indicate some of the sites where Sgf11 and Pbp45 co-localise in the loci, corresponding to snRNA genes (34AB, 96A, 95C, 23A etc.). Arrowheads indicate some of the loci where Sgf11 and Pbp45 do not co-localise (82E, 60F). Each site is indicated according the D. melanogaster Chromosome Map (FlyBase.org). The enlarged fragments of merged image (shown in frames) are presented on right panels. Scale bar = 10μm.
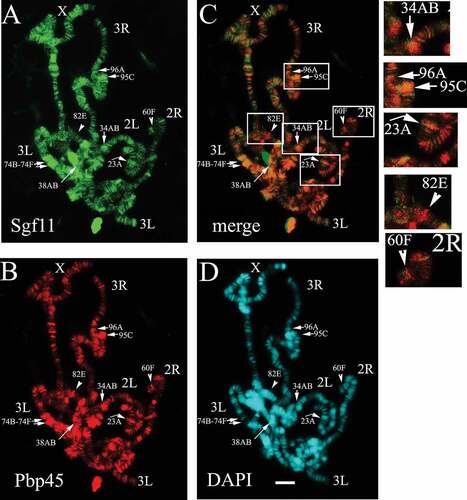
The double immunostaining of polytene chromosomes from the salivary glands of Drosophila using antibodies against Sgf11 () and Pbp45 () was carried out in accordance with [Citation30,Citation31]. It has been previously shown that the main pool of Drosophila Sgf11 is associated with DUB module of SAGA, and Sgf11 is present at a substantial number of loci on the polytene chromosomes of Drosophila [Citation29]. The Pbp45 protein was also detected at many loci which suggested an extensive involvement of this protein in the regulation of gene transcription (). Although in Drosophila, it is not clear whether Pbp45 is involved in transcription of other genes, apart from the snRNA genes, its human homolog, hSNAPC1, was shown to be present at the genes other than snRNA [Citation32]. These data are in good agreement with our results demonstrating that Pbp45 binds multiple sites on the Drosophila polytene chromosomes.
Pbp45 and Sgf11 colocalise at many actively transcribed genes on polytene chromosomes (34AB, 96A,C, 23A, etc.). At the same time, these factors were detected at many sites (82E, 60F, etc.) independently from each other (). What is significant is that the two proteins, Sgf11 and Pbp45, colocalise at those sites where U1 – U6 snRNA genes are located ().
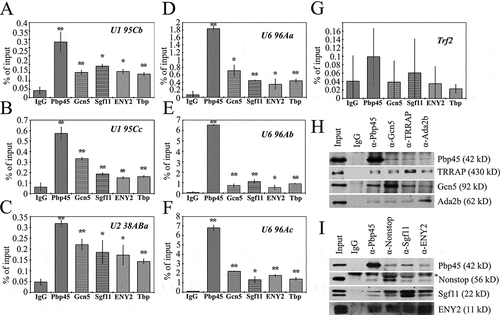
To confirm that SAGA complex is indeed present at the promoter regions of the snRNA genes, we performed chromatin immunoprecipitation (CHIP) with antibodies against the components of the SAGA (Gcn5, ENY2, and Sgf11) and Pbp45 (). For each studied snRNA gene, we designed specific primers which allowed PCR amplification of its promoter regions. Additionally, to roughly estimate the transcription levels of each individual gene, we verified the level of TBP on its promoter. As a negative control, a fragment of the coding region of the trf2 gene, encoding transcription factor trf2 (the TBP related factor 2) was used. Our results demonstrated that none of factors studied here bind to trf2 (). As expected, Pbp45 was present at the promoters of all snRNA genes (). All the SAGA complex proteins chosen for the analysis were detected at the promoter regions of snRNA genes, both at promoters of the genes transcribed by RNA polymerase II (snRNA U1 and snRNA U2) and at promoters of the U6 snRNA genes which are transcribed by RNA polymerase III. The amounts of the analyzed factors at the promoters to a large extent correlated with the transcription levels of corresponding genes, which could be roughly estimated by the presence of TBP at the promoters. The presence of ENY2 and Sgf11, the SAGA DUB module components, shows that SAGA participates in the regulation of transcription of the studied genes.
Figure 2. SAGA is recruited to snRNA and is associated with Pbp45. (a-i) Occupation of the U promoters (U1 95Cb, U1 95Cc, U2 38Aba, U6 96Aa, U6 96Ab, U6 96Ac) and the fragment of the coding region of trf2 gene (control, see Supplementary, Table 1) by Pbp45, ENY2, Sgf11, Gcn5, and TBP. The protein level was measured by ChIP. The levels of studied proteins on the trf2 fragment (control) are extremely low (g). The results of ChIP are provided as a percentage of input (here, and in ). Here and on the figures below the graphs show means +- S.E.M. of three independent experiments. t-tests have been performed to compare the means (*P < 0.05, and **P < 0.01). (h, i) – Drosophila S2 cell nuclear extract treated with DNase and RNase was immunoprecipitated with affinity purified polyclonal antibodies against Pbp45 and HAT module and TRRAP of SAGA components (h), or Pbp45 and DUB module of SAGA components (i). 1/5 of the Input, and equal amounts of IP precipitates (1/5) were loaded per each lane. Protein sizes were calculated based on the concurrently run protein size markers. Rabbit Immunoglobulin G (IgG) coupled to protein A-Sepharose beads was used as a control. Equivalent amounts of the input fraction, and of the proteins bound to the immunosorbent (IP) were separated by SDS-PAGE and analyzed by western blotting with the above-mentioned antibodies. Asterisk indicates antibody chains.
To determine whether the proteins in the SAGA complex physically interact with the snRNA gene transcription apparatus, we co-immunoprecipitated the Pbp45 protein with the components of the two SAGA modules, the HAT module (Gcn5 and Ada2b factors), TRRAP, and the DUB module (Sgf11, ENY2, and Nonstop) from the nuclear extract of the Drosophila S2 cells. Co-immunoprecepitation results (,); Supplementary Figure 2(a-b)) demonstrated that the protein components of both the HAT and DUB modules interact with Pbp45, and thus, with the PBP, the component of the snRNA transcription apparatus.
Therefore, the SAGA complex proteins are not only present at the promoter regions of snRNA genes, but they also interact with the snRNA transcription machinery. Moreover, SAGA participates in snRNA gene transcription by both Pol II and Pol III.
The presence of SAGA at the RNA polymerase III target genes and its interaction with Brf1
To study the interaction between SAGA and the RNA polymerase III transcription apparatus, we produced rabbit polyclonal antibodies against the Brf1 protein (Supplementary Figure 1(b)), the component of the RNA polymerase III basal transcription complex [Citation33]. The antibodies were affinity purified and their specificity was verified by RNAi knockdown of Brf1 (Supplementary Figure1(d)).
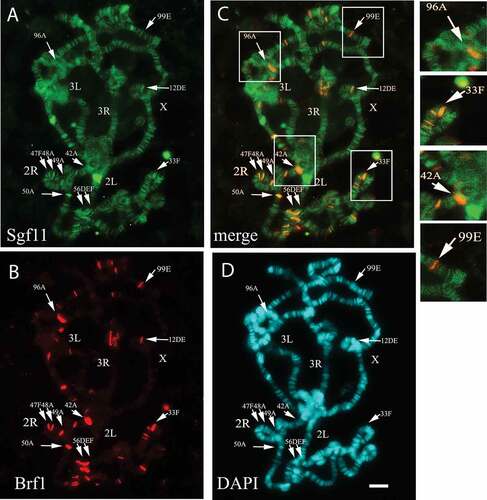
To detect SAGA at the RNA polymerase III-transcribed genes, we analyzed colocalization of Sgf11 () and Brf1 () on polytene chromosomes. In contrast to Sgf11, which is present at many active transcription sites in interbands and puffs, Brf1 was found in a relatively low number of sites which corresponded to localization of the Pol III-transcribed genes (). However, the immunostaining experiments revealed that Sgf11 was present at all Brf1 sites. In particular, they colocalise at the loci corresponding to U6 and tRNA genes, as well as other RNA polymerase III target genes ().
Figure 3. SAGA is present at all Brf1 sites on Drosophila polytene chromosomes. (a) Drosophila polytene chromosomes stained with anti-Sgf11 antibodies, (b) with anti-Brf1 antibodies, and (c) merged images. (d) Chromosomes were co-stained with DAPI. The Drosophila chromosomes (X, 2L, 2R, 3L and 3R) are indicated. Arrows indicate the sites where Sgf11 and Brf1 co-localise. Each site is indicated according the D. melanogaster Chromosome Map (FlyBase.org). The enlarged fragments of merged image (shown in frames) represented on right panel demonstrate the colocalisation of Sgf11 and Brf1 at loci containing U6 RNA and tRNA genes: 96A (U6 RNA), 33F (U6 and tRNA), 42A (tRNA), 99E (tRNA). Scale bar = 10μm.
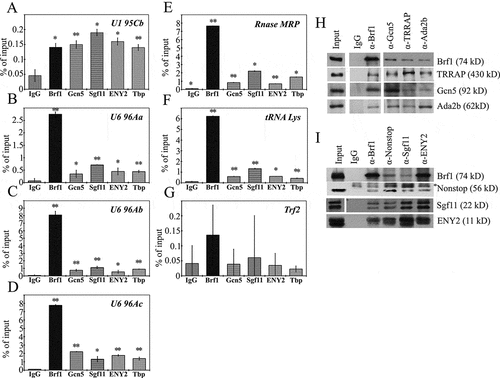
Chromatin immunoprecipitation using antibodies against the components of the SAGA complex and Brf1 confirmed that SAGA is present not only at the promoters of individual U6 snRNA genes (), but also at the promoters of other genes (Rnase MRP and tRNA Lys) transcribed by RNA polymerase III ().The trf2 gene fragment which does not to bind with SAGA [Citation34] was used as a control.
The physical interaction between the SAGA proteins and the RNA polymerase III transcription apparatus was tested by co-immunoprecipitation using antibodies against proteins of the SAGA complex HAT module (Gcn5 and Ada2b), TRRAP, or the DUB module (Sgf11, ENY2, and Nonstop), as well as antibodies against the Brf1 protein (), Supplementary Figure 2(a,b)). We have demonstrated that the protein components of both the HAT module and the DUB module interact with Brf1. All the tested SAGA subunits interacted with Brf1 in the nuclear extract. It is worth noting, however, that the strongest interaction was observed between Sgf11 and Brf1. The anti-Sgf11 antibodies almost completely depleted Brf1 from the nuclear extract (Supplementary Figure 2(c), compare Outputs for IgG, anti-Brf1, and anti-Sgf11 antibodies). Other DUB module subunits showed weaker interaction with Brf1, which possibly implies that Sgf11 directly interacts with Brf1. It should be also pointed out that a complete depletion of Brf1 from the nuclear extract by anti-Sgf11 antibodies indicates that Brf1 interacts with Sgf11 (or DUB module) at all target genes, and not only at the genes tested here (), this finding is in good agreement with the polytene chromosome immunostaining results ().
Figure 4. SAGA is recruited to Pol III-dependent genes and is associated with Brf1. (a-i) Occupation of the U6 promoters (U6 96Aa, U6 96Ab, U6 96Ac) and promoters of other Pol III dependent genes (Rnase MRP and tRNA Lys) by Brf1, TBP, and the SAGA components (ENY2, Sgf11, and Gcn5). The trf2 gene fragment (see Supplementary, ) was used as a control. The protein levels were measured by ChIP. The results of ChIP are provided as a percentage of input. Presented data for SAGA subunits Sgf11, Gcn5, and Eny2 are of the same ChIP experiment as in . The statistical tests for this figure are the same as described in the legend to . (h) Drosophila S2 cells nuclear extract treated with DNase and RNase was immunoprecipitated with affinity purified polyclonal antibodies against Brf1 and subunits of SAGA HAT module, and TRRAP (indicated at the top). (i) The same extract immunoprecipitated with affinity purified polyclonal antibodies against Brf1 and subunits of SAGA DUB (indicated at the top). 1/5 of the Input, and equal amounts of IP precipitates (1/3 of Input) were loaded per each lane. Rabbit Immunoglobulin G (IgG) coupled to protein A-Sepharose beads was used as a control in (H and I). Equivalent amounts of the input fraction, and of the proteins bound to the immunosorbent (IP) were separated by SDS-PAGE and analyzed by western blotting with the above-mentioned antibodies. Asterisk indicates antibody chains.
Therefore, SAGA complex is present at the promoters of the RNA polymerase III-transcribed genes and interacts with the RNA polymerase III transcription apparatus.
The effect of mutations in the genes encoding SAGA complex subunits on snRNA transcription
Next, we addressed the question, whether specific impairing of individual subunits of the SAGA complex could affect the snRNA gene transcription in flies. The effect of mutations in genes encoding the SAGA subunits on the snRNA transcription in Drosophila showed that it is indeed the case. First, we confirmed that all studied factors were present on the promoters of snRNA genes in flies () (chromatin was prepared from Drosophila pupae), as it was shown in the previous section for snRNA genes in the S2 cells. So, SAGA also participates in the snRNA gene regulation at the level of the whole organism.
Figure 5. The participation of SAGA complex in snRNA transcription. (a-c) Occupation of the U1 and U6 promoters (U1 95Cc, U6 96Ac) and trf2 fragment (control) by Brf1, Pbp45, Gcn5, Sgf11, ENY2, and TBP in Drosophila pupat. The protein level was measured by ChIP. The results of ChIP are provided as a percentage of input. (d) – Effects of decrease of Ada2b, Gcn5, Sgf11, and Nonstop transcription levels in mutant strains (shown in Supplementary. ), on the level of transcription of U1, U6, tRNA Lys and 7SK genes. TM6B, Tb[Citation1]/+ strain was used as a control. The transcription levels were normalised on 28S rRNA. (e) Effects of decrease of Ada2b, Gcn5, Sgf11, and Nonstop transcription levels in mutant strains (shown in Suppl. ), on the level of Brf1 on promoters of U1 and U6 genes. (f) The effect of mutations of the SAGA subunits on H2B monoubiquitylation of U6 96 Ac promoter. The levels of nonmodified and monoubiquitylated H2B were measured by ChIP in the control and mutant strains (indicated below the Figure). The antibodies against nonmodified H2B and against monoubiquitylated H2B (H2BK120Ub) were used. The data represent the ratio of the H2BK120Ub level to the level of nonmodified H2B. The statistical tests for this figure are the same as described in the legend to .
![Figure 5. The participation of SAGA complex in snRNA transcription. (a-c) Occupation of the U1 and U6 promoters (U1 95Cc, U6 96Ac) and trf2 fragment (control) by Brf1, Pbp45, Gcn5, Sgf11, ENY2, and TBP in Drosophila pupat. The protein level was measured by ChIP. The results of ChIP are provided as a percentage of input. (d) – Effects of decrease of Ada2b, Gcn5, Sgf11, and Nonstop transcription levels in mutant strains (shown in Supplementary. Figure 3(a)), on the level of transcription of U1, U6, tRNA Lys and 7SK genes. TM6B, Tb[Citation1]/+ strain was used as a control. The transcription levels were normalised on 28S rRNA. (e) Effects of decrease of Ada2b, Gcn5, Sgf11, and Nonstop transcription levels in mutant strains (shown in Suppl. Figure 3A), on the level of Brf1 on promoters of U1 and U6 genes. (f) The effect of mutations of the SAGA subunits on H2B monoubiquitylation of U6 96 Ac promoter. The levels of nonmodified and monoubiquitylated H2B were measured by ChIP in the control and mutant strains (indicated below the Figure). The antibodies against nonmodified H2B and against monoubiquitylated H2B (H2BK120Ub) were used. The data represent the ratio of the H2BK120Ub level to the level of nonmodified H2B. The statistical tests for this figure are the same as described in the legend to Figure 2.](/cms/asset/d25eaaa4-d18f-4d0f-b5c9-ae4525538c13/kccy_a_1489175_f0005_b.gif)
The studied mutant strains included mutations in genes encoding subunits of the DUB module (Sgf11 and Nonstop) and the HAT module (Gcn5 and Ada2b). Each mutation caused a complete elimination of the corresponding protein and was lethal in homozygotes (Supplementary Table 1). Thus, we performed our experiments on heterozygous strains. However, in all strains, the level of corresponding mutant gene expression was decreased (Supplementary Figure 3(a)). The TM6,Tb[Citation1]/+ heterozygotes were used as a control. The flies with mutations in the studied genes, as well as the control flies, were used to isolate RNA to measure the transcription level of the studied genes by qRT-PCR.
The snRNA (U1 and U6) transcription level in the mutant flies was compared with that in the control strain. We also measured the transcription levels of two other Pol III dependent genes (tRNA Lys and 7SK). We observed that the mutations in all the tested SAGA complex subunit genes caused decrease in the U1 and U6 snRNA transcription levels with a less significant effect on the U1 snRNA transcription (). Therefore, mutations in the genes encoding SAGA subunits cause a decrease in transcription of snRNAs, as well as of Pol III-dependent genes, thus confirming that SAGA is indeed the coactivator of their transcription. However, the effect of SAGA on the Pol III-dependent snRNA transcription is stronger than on the Pol II-dependent snRNA transcription. Decreased transcription levels were also observed for tRNA Lys and 7SK ().
The SAGA complex is the transcriptional coactivator for a large number of genes. We show that mutations in SAGA subunits did not have significant effect on the transcription levels of the other factors which participate in the transcription of snRNA and Pol III-dependent genes and that SAGA directly regulates the snRNA and Pol III-dependent genes (Supplementary Figure3).
Finally, we investigated if mutations in SAGA subunits affected the attachment of Brf1 to promoters of U6 genes (). As expected, no Brf1 was found on the promoter of U1, used as a control. The high level of Brf1 found on U6 promoters, was significantly decreased in the mutated fly strains. These results indicate that SAGA may be also involved in the recruitment of Pol III transcription apparatus.
The level of H2B monoubiquitylation was also affected by SAGA mutations (). We investigated the H2B monoubiquitylation of U6 96Ac, since this gene was transcribed more actively than the other studied genes, as was roughly estimated using the TBP level on its promoter as a proxy (). The higher transcription level allowed us to get more prominent results, and better estimate the effect of mutation on H2B monoubiquitylation. The antibodies against nonmodified H2B and against monoubiquitylated H2B (H2BK120Ub) were used. The data () represent the ratio of H2BK120Ub to nonmodified H2B, verified by ChIP in the control and mutant strains. These data demonstrate that the H2B monoubiquitylation increased in all mutated flies comparatively to the control.
Discussion
The current work investigates the role of the SAGA complex in snRNA transcription in Drosophila. We have demonstrated that SAGA complex is present at the promoters of snRNA genes and the subunits of the SAGA complex interact with PBP, the component of the snRNA transcription apparatus. We have shown that SAGA coactivates transcription of both the Pol II and Pol III-dependent snRNA genes. Moreover, we have detected SAGA at the promoters of several Pol III-dependent genes (Rnase MRP and tRNA Lys) and found an interaction between the SAGA subunits and Brf1, one of the key proteins within the RNA polymerase III transcription machinery. Decrease of the SAGA complex subunits levels in mutant flies caused the reduction of the U1 and U6 snRNA transcription levels.
We examined the presence of the factors within the SAGA complex at the small nuclear RNA genes. The study of Sgf11 and Pbp45 colocalization on polytene chromosomes revealed that Pbp45 can be found not only at the snRNA loci but also at other actively transcribed sites (in puffs and interbands). Similar data were obtained previously by the whole genome sequencing of hSNAPC1, the human Pbp45 homolog [Citation32]. Our findings are the first indication that similar to hSNAPC1, dPbp45 is associated in addition to the snRNA gene localization sites, with other sites of active transcription by Pol II.
It has been demonstrated earlier that the SAGA complex components, TRRAP and Gcn5, are involved in the transcription of the RNA Pol III- dependent human genes [Citation25]. It was demonstrated that these proteins have effect on the tRNA and 5S rRNA transcription, which was characterised by the acetylation of histone H3, but not the histone H4, similar to what is observed for the transcription of the Pol II-dependent genes. This work has demonstrated that TRRAP and Gcn5 are recruited to Pol III by c-Myc, although a certain portion of these factors were recruited in a c-Myc-independent manner [Citation25]. Recently, a ChIP-seq analysis of Drosophila embryos has shown that Ada2b, Nonstop, and Sgf11 occupied promoter regions of snRNA [Citation26]. Our results obtained for Drosophila, support these data and we have demonstrated that SAGA participates in the transcription of snRNAs. We have also shown that the SAGA complex participates in the transcription of other Pol III target genes, namely the U6 snRNA genes.
Our genetic analysis and co-immunoprecipitatin experiments revealed that the DUB module has a substantial effect on the transcriptional activation of U6 snRNA genes. A strong interaction between the Sgf11 subunit of the DUB module and the Brf1 subunit of the Pol III transcription apparatus suggests that DUB (probably, as a part of the SAGA complex) may play a role of transcriptional coactivator for at least a substantial pool of Pol III-transcribed genes. These results are in agreement with the data that DUB module has both SAGA-related and independent functions [Citation26].
Furthermore, the interaction between Sgf11 and Brf1, the Pol III transcription apparatus subunit, was so strong that it could be supposed that Sgf11 may exert some functions in the mode independent from the SAGA complex. It might be possible as well that at these snRNA genes, the DUB subcomplex deubiquitinates not only the histone H2B, but also some other components of the Pol III transcriptional machinery. Sgf11 also proved to interact with Brf1 more strongly than other SAGA subunits. These findings are indicative of Sgf11 playing some specific part in this interaction which is not related to the SAGA complex function.
In summary, we have demonstrated that the SAGA complex factors are present at the promoters of the snRNA genes, interact with the Pol II and Pol III-dependent snRNA gene transcription apparatus, and participate in the transcriptional coactivation of these genes.
Materials and methods
Fly strains
Mutant fly strains: Sgf11 (RRID:BDSC_17941: w1118; PBac{RB}[Citation10]Sgf11e01308/TM6B, Tb[Citation1]), Nonstop (RRID:BDSC_11553: P{ry[+ t7.2] = PZ}not[02069] ry[506]/TM6B, ry[CB] Tb[+])and Gcn5 (RRID:BDSC_18868: w[1118]; PBac{w[+ mC] = WH}Gcn5[f05456]/TM6B, Tb[Citation1]) were obtained from the Bloomington Stock Center (http://flystocks.bio.indiana.edu/). Ada2b[d842]/TM6C, Tb[Citation1],Sb[Citation1] strain was kindly provided by I. M. Boros [Citation35]. TM6B, Tb[Citation1]/+ flies were used as a control. More information about fly strains is provided in [Citation13,Citation35,Citation36].
Antibodies
Polyclonal antibodies against Pbp45 (aa 186–363), Brf1 (aa 492–662) were obtained in our laboratory; rabbits were immunised with the corresponding His6-tagged protein fragments. Affinity purified antibodies were used in all experiments. Antibodies against ENY2, Sgf11, TBP, Gcn5, Ada2b, Nonstop, and TRRAP were described previously [Citation29–Citation31,Citation34,Citation37]. Antibodies against histone H2B (17–10,054), histone Ubiquityl-H2B (17–650) were purchased in Millipore.
Immunostaining
Immunostaining of polytene chromosomes was performed as described previously [Citation29,Citation30] using mouse anti-Sgf11, rabbit anti-Pbp45, and anti-Brf1, and corresponding secondary antibodies (Molecular Probes). The results were analyzed using DMR/HC5 fluorescence microscope (Leica) with an HCX PZ Fluotar 100/1.3 objective lens and recorded using a Leica DC350 F digital camera.
Drosophila cell culture extracts.
Drosophila S2 cells were maintained in Schneider’s insect medium (Sigma) containing 10% fetal bovine serum (HyClone, United States) at 25°C. To extract proteins, S2 cells were lyzed. Cells were centrifuged at 2000 rpm at + 4°C for 5 min and resuspended in 1 ml of 1x PBS with 25x PIC (Protease Inhibitor Cocktail, Roche). Cells were then centrifuged once more at 2000 rpm for 5 min at 4°C and resuspended in LB buffer, containing 10 mM Hepes pH 7.0, 0.4 M NaCl, 5 mM MgCl2, 0.5% NP-40, 25x PIC, 1 mM DTT, and 0.3 μL of DNAse I, at the ratio of 1/10. Cells were incubated in LB buffer for 20 min on ice and the lysate was further centrifuged at 13,000 rpm for 15 min at 4°C.
Immunoprecipitation
Antibodies were bound to protein A Sepharose (Pharmacia) according to the manufacturer’s recommendations. 100 μL of the protein extract in IP buffer (10 mМ HEPES, рН 7.9, 5 mМ MgCl2, 0.1% NP-40, 0.15 М NaCl, 25x PIC, 1 mM DTT, RNase (Stratagene, 10 U/mL), and 0.5 ng/μL of DNAse I) were incubated with 10 μL of Sepharose at 4°C overnight. Sepharose was then sedimented, supernatant was discarded, and the precipitate was washed three times with 100 μL of IP500 buffer (10 mМ HEPES, рН 7.9, 5 mM MgCl2, 10% glycerol, 500 mM KCl, and 0.1% NP-40) for 10 min each. An equal volume of the electrophoresis Laemmli loading buffer was further added to the precipitate, the obtained suspension was boiled, centrifuged, and supernatant was applied to the polyacrylamide gel for subsequent Western blotting.
Chromatin immunoprecipitation
Chromatin for a single chromatin immunoprecipitation experiment was prepared from 3 × 10 [Citation6] Schneider S2 cells or Drosophila pupae, or flies Oregon-R. Cross-linking was performed by adding formaldehyde (final concentration 1.5%) to the incubation medium for 15 min at room temperature and stopped by the addition of 2.5M glycine (1/20 of the total volume). After washing with three volumes of cold (4°C) PBS, chromatin was resuspended in 0.2 mL of SDS-containing buffer (50 mM HEPES KOH, pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, and 0.1% SDS) with protease inhibitors cocktail (Roche) and sheared to 500-bp fragments by sonication. Sonicated chromatin was centrifuged twice at 16 000 g for 20 min, and used in immunoprecipitation experiments. About 10 µg of antibodies and 15 µL of Protein A Sepharose (Sigma) were taken for a single experiment; ssDNA and BSA were added to a final concentration of 1 mg/mL. The precipitated chromatin was sequentially washed with SDS-containing buffer, SDS-containing buffer with 0.5 M NaCl, LiCl-containing buffer (20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, and 0.5% Na deoxycholate), and TE buffer (20 mM Tris-HCl, pH 8.0 and 1mM EDTA). Precipitated chromatin complexes were eluted by two sequential incubations in the elution buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 1% SDS), 30 min each, at room temperature. 5 M NaCl was added to the eluted chromatin solution (16 µL per 500 µL sample) and incubated at 65°C for 16 h in thermoshaker for de-crosslinking. De-crosslinked chromatin was treated with proteinase (3 µL of proteinase K and 5 µL of 0.5 M EDTA per 500 µL sample) for 4 h at 55°C in thermoshaker. DNA was extracted with a phenol/chloroform mixture and precipitated with isopropanol. The precipitate was dissolved in TE buffer and analyzed by real-time PCR (qPCR). The results of immunoprecipitation experiments were expressed as the percent ratio of the precipitated chromatin fraction to the input chromatin fraction. The sequences of the primers used are available on request. Each measurement was carried out in at least five repeats, and the mean value was calculated.
RNA isolation
Total RNA was extracted from Drosophila imago with Trizol (Invitrogen) according to the manufacturer’s recommendations. cDNA was produced by RevertAid Reverse Transcriptase (Thermo Scientific Inc., USA). Reverse transcription was performed with the Random primer. The levels of mRNAs were measured by qPCR using pairs of primers described in Supplemental .
Table 1. Mutant fly strains.
Statistical analysis
Comparisons between the experimental groups were made using the Student’s t-test. All data are presented as means ± SD (error bars) for at least three independent experiments. Asterisks indicate that the data are statistically significant at *P < 0.05 or **P < 0.01.
Supplemental Material
Download Zip (11.4 MB)Acknowledgments
The authors are grateful to Vladic Mogila for his help in preparing the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplementary data can be accessed here
Additional information
Funding
References
- Jawdekar GW, Henry RW. Transcriptional regulation of human small nuclear RNA genes. Biochim Biophys Acta. 2008;1779:295–305.
- Zieve G, Benecke BJ, Penman S. Synthesis of two classes of small RNA species in vivo and in vitro. Biochemistry. 1977;16:4520–4525.
- Lobo SM, Lister J, Sullivan ML, et al. The cloned RNA polymerase II transcription factor IID selects RNA polymerase III to transcribe the human U6 gene in vitro. Genes Dev. 1991;5:1477–1489.
- Baer M, Nilsen TW, Costigan C, et al. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990;18:97–103.
- Hung KH, Stumph WE. Regulation of snRNA gene expression by the Drosophila melanogaster small nuclear RNA activating protein complex (DmSNAPc). Crit Rev Biochem Mol Biol. 2011;46:11–26.
- Zamrod Z, Tyree CM, Song Y, et al. In vitro transcription of a Drosophila U1 small nuclear RNA gene requires TATA box-binding protein and two proximal cis-acting elements with stringent spacing requirements. Mol Cell Biol. 1993;13:5918–5927.
- Dahlberg JE, Lund E. Structure and expression of U-snRNA genes. Mol Biol Rep. 1987;12:139–143.
- Henry RW, Mittal V, Ma B, et al. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 1998;12:2664–2672.
- Lai H-T, Kang YS, Stumph WE. Subunit stoichiometry of the Drosophila melanogaster small nuclear RNA activating protein complex (SNAPc). FEBS letters. 2008;582:3734–3738.
- Guiro J, Murphy S. Regulation of expression of human RNA polymerase II-transcribed snRNA genes. Open Biol. 2017;7.
- Canella D, Praz V, Reina JH, et al. Defining the RNA polymerase III transcriptome: genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710–721.
- Koutelou E, Hirsch CL, Dent SY. Multiple faces of the SAGA complex. Curr Opin Cell Biol. 2010;22:374–382.
- Weake VM, Lee KK, Guelman S, et al. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405.
- Weake VM, Swanson SK, Mushegian A, et al. A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev. 2009;23:2818–2823.
- Zhao Y, Lang G, Ito S, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101.
- Gurskii D, Nabirochkina EN, Kopytova DV. [The role of multifunctional coactivator complex saga in regulation of eukaryotic gene expression]. Mol Biol (Mosk). 2013;47:914–921.
- Martinez E, Palhan VB, Tjernberg A, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795.
- Spedale G, Timmers HT, Pijnappel WW. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012;26:527–541.
- Samara NL, Datta AB, Berndsen CE, et al. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science (New York, NY). 2010;328:1025–1029.
- Kuo MH, Brownell JE, Sobel RE, et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272.
- Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133.
- Nagy Z, Riss A, Fujiyama S, et al. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell Mol Life Sci. 2010;67:611–628.
- Guelman S, Suganuma T, Florens L, et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 2006;26:871–882.
- Helmlinger D, Tora L. Sharing the SAGA. Trends Biochem Sci. 2017;42:850–861.
- Kenneth NS, Ramsbottom BA, Gomez-Roman N, et al. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc Natl Acad Sci U S A. 2007;104:14917–14922.
- Li X, Seidel CW, Szerszen LT, et al. Enzymatic modules of the SAGA chromatin-modifying complex play distinct roles in Drosophila gene expression and development. Genes Dev. 2017;31:1588–1600.
- Chen J, Ezzeddine N, Waltenspiel B, et al. An RNAi screen identifies additional members of the Drosophila Integrator complex and a requirement for cyclin C/Cdk8 in snRNA 3’-end formation. RNA (New York, NY). 2012;18:2148–2156.
- Takahashi H, Takigawa I, Watanabe M, et al. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat Commun. 2015;6:5941.
- Gurskiy D, Orlova A, Vorobyeva N, et al. The DUBm subunit Sgf11 is required for mRNA export and interacts with Cbp80 in Drosophila. Nucleic Acids Res. 2012;40:10689–10700.
- Soldatov A, Nabirochkina E, Georgieva S, et al. TAFII40 protein is encoded by the e(y)1 gene: biological consequences of mutations. Mol Cell Biol. 1999;19:3769–3778.
- Vorobyeva NE, Soshnikova NV, Nikolenko JV, et al. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc Natl Acad Sci U S A. 2009;106:11049–11054.
- Baillat D, Gardini A, Cesaroni M, et al. Requirement for SNAPC1 in transcriptional responsiveness to diverse extracellular signals. Mol Cell Biol. 2012;32:4642–4650.
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620.
- Kurshakova MM, Krasnov AN, Kopytova DV, et al. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007;26:4956–4965.
- Pankotai T, Komonyi O, Bodai L, et al. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol Cell Biol. 2005;25:8215–8227.
- Poeck B, Fischer S, Gunning D, et al. Glial cells mediate target layer selection of retinal axons in the developing visual system of Drosophila. Neuron. 2001;29:99–113.
- Muratoglu S, Georgieva S, Papai G, et al. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol Cell Biol. 2003;23:306–321.
