ABSTRACT
The proteasome is the key player in targeted degradation of cellular proteins and serves as a therapeutic target for treating several blood malignancies. Although in general, degradation of proteins via the proteasome requires their ubiquitination, a subset of proteins can be degraded independently of their ubiquitination by direct interaction with subunits of the 20S proteasome core. Thus, investigation of the proteasome-associated proteins may help identify novel targets of proteasome degradation and provide important insights into the mechanisms of malignant cell proteostasis. Here, using biochemical purification of proteasomes from multiple myeloma (MM) cells followed by mass-spectrometry we have uncovered 77 proteins in total that specifically interacted with the 20S proteasome via its PSMA3 subunit. Our GST pull-down assays followed by western blots validated the interactions identified by mass-spectrometry. Eleven proteins were confirmed to bind PSMA3 only upon apoptotic conditions induced by a combined treatment with the proteasome inhibitor, bortezomib, and genotoxic drug, doxorubicin. Nine of these eleven proteins contained bioinformatically predicted intrinsically disordered regions thus making them susceptible to ubiquitin-independent degradation. Importantly, among those proteins five interacted with the ubiquitin binding affinity matrix suggesting that these proteins may also be ubiquitinylated and hence degraded via the ubiquitin-dependent pathway. Collectively, these PSMA3-interacting proteins represent novel potential substrates for 20S proteasomes upon apoptosis. Furthermore, these data may shed light on the molecular mechanisms of cellular response to chemotherapy.
Abbreviations: BD: bortezomib/doxorubicin treatment; CDK: cyclin-dependent kinases; CHCA: α-cyanohydroxycinnamic acid; IDP: intrinsically disordered proteins; IDR: intrinsically disordered regions; IPG: immobilized pI gradient; MALDI TOF/TOF: matrix-assisted laser desorption/ionization time-of-flight tandem mass-spectrometry; MM: multiple myeloma; ODC: ornithine decarboxylase; PI: proteasomal inhibitors; PSMA: alpha-type 20S proteasome subunits; PTMs: post-translational modifications; SDS-PAGE: sodium dodecylsulphate polyacrylamide gel electrophoresis; UIP: ubiquitin-independent proteasomal proteolysis.
Introduction
The 26S proteasome is a multi-subunit ribonucleoprotein complex found in both nuclei and cytoplasm of all eukaryotic cells. It consists of a barrel-shaped 20S core flanked by either one or two regulatory 19S complexes. The main function of cellular proteasomes is the cleavage of proteins labeled with polyubiquitin chains, through limited and controlled proteolysis. All three proteolytic activities of the proteasome (caspase-, trypsin-, and chymotrypsin-like) are associated with beta subunits of the 20S core (β1, β2, β5). On the contrary, the alpha-type subunits are shown to play structural role [Citation1]. Recently, we and other groups have shown that alpha-subunits possess with endo-ribonuclease activity thereby controlling expression levels of several important transcription factors [Citation2]. Furthermore, proteasomes were shown to associate with RNA. The RNA component of proteasomes is represented by a heterogeneous fraction of nucleic acid molecules ranging from 20 to 120 nucleotides, recently shown to also contain miRNAs [Citation3].
The regulatory complexes participate in the substrate recognition and unfolding and control the size of proteolysis products. The data accumulated over the past decades point to 26S proteasomes as universal intracellular instruments carrying an integrated set of diverse biochemical functions. Although mostly recruited as a whole, a proteasome is able to utilize one or more of its functions depending on a set of specific post-translational modifications [Citation4].
An ever-increasing number of studies suggests that certain intracellular proteins can undergo proteasome-mediated proteolysis without the ubiquitin mark involved. In that case, the specificity of degradation is mediated by binding between the proteasome and the substrate protein via additional interactions with accessory proteins. Furthermore, certain amino acids motifs for proteasome-specific degradation must be intrinsically present in the substrate protein itself. To expose such signals to the catalytic subunits of 20S complexes the substrate proteins are unfolded using ATPase activity of 19S proteasomes. Alternatively, the substrate proteins can be intrinsically disordered to provide such an access to beta-subunits. Noteworthy, approximately 20% of all cellular proteins can be degraded in vitro by the 20S proteasome directly due to the presence of extended disordered regions in their sequences [Citation5]. However, this large number of proteins cannot undergo spontaneous unregulated degradation in the living cell implying that some level of targeting specificity takes place. This is likely achieved through the interaction between the proteasome and its substrate proteins. To this end, several proteins including ornithine decarboxylase (ODC) and the IκBα subunit of the NF-kB transcription factor were shown to undergo specific degradation by the 20S proteasome [Citation6]. Furthermore, the major tumor suppressor in vertebrates, p53, was also demonstrated to undergo ubiquitin-independent degradation [Citation7]. Importantly, since p53 and related proteins, p73 and p63, affect expression of many genes not only directly, but also indirectly, via regulation of micro-RNAs, studying the mechanisms of proteasome-mediated degradation of these proteins is still topical [Citation8,Citation9]. A similar mechanism of degradation was shown for an important CDK inhibitor p21WAF1/CIP1. The latter contains the degradation sequence located at the carboxyl terminus of the protein that interacts with PSMA3 subunit of the 20S proteasome core [Citation10,Citation11]. pRb and EGR-1 also interact with PSMA3, which apparently results in their degradation [Citation12,and Citation13, respectively]. Therefore, PSMA3 may be considered as a gateway to the 20S proteasome core. Taking into consideration that all these substrates participate in cancer-related signaling pathways, the identification of PSMA3-associated proteins may be important for understanding of aberrations in the complex networking during the tumor development.
Because the proteasome is considered as a key player in variety of cellular regulatory pathways [Citation14,Citation15], inhibition of its activity draws attention as a promising therapeutic target for treating various cancers. Multiple myeloma (MM) is a tumor specific to plasma cells. The characteristic feature of these cells is production of large amounts of abnormal antibodies. Although the disease is considered as incurable, admission of proteasomal inhibitors (PI) and PI-based combination therapies resulted in temporary remission of patients. The molecular mechanism for this phenomenon is several fold: first, multiple myeloma cells are more susceptible to proteasome inhibitors when compared to normal plasma cells [Citation16]. Second, highest efficacy of proteasome inhibitors is achieved for the cells exhibiting high rates of protein synthesis. This means that the tumor cells experience constant proteotoxic stress and are therefore more sensitive to the inhibition of proteasomes. Accordingly, when the proteasome activity is blunted in MM cells that actively produce abnormal antibodies, malfunctioning proteins may start to accumulate inside the cell, with a high chance of activation of cellular unfolded protein response that eventually triggers apoptosis [Citation17].
With that in mind, the proteasomal activity appears instrumental for MM cell survival and hence, for the outcome of therapeutic treatment. Importantly, we have shown previously that genotoxic stress attenuates the activity of proteasomes via multiple post-translational modifications, including phosphorylation [Citation4]. Thus, the detailed characterization of proteasome-related interactome under genotoxic stress conditions may assist in better understanding of the chemotherapy-related molecular events that guide MM cells to apoptosis. Finally, revealing the proteins that bind to the 20S proteasome core may uncover new adapter proteins that target biologically significant regulators for degradation. The latter may additionally broaden our knowledge of the protein utilization pathways in cancer cells.
Results
Identification of PSMA3-interacting proteins by GST pull-down approach and proteomics. The aim of this work was to conduct the comparative investigation of the repertoire of proteins associated with proteasomes in MM cells upon the conditions of normal growth and apoptosis induced with genotoxic drug, doxorubicin. To enrich the population of the labile proteins after genotoxic stress we treated the RPMI 8226 cells with doxorubicin simultaneously with bortezomib (BD), which specifically inhibits trypsin- and chymotrypsin activities of the proteasome (). Separate analysis of the cytoplasmic and nuclear fractions allowed us to distinguish the differences in protein composition in the cytoplasm and nucleus of MM cells upon genotoxic stress. In fact, we observed a drastic difference in protein expression patterns in the cytoplasm samples of B/D-treated versus non-treated cells several (, panel C). Importantly, the concentrations of drugs that we used in this study were selected to specifically induce apoptosis and avoid necrosis of the cells. The effects of different concentrations of drugs on cell death were monitored by flow cytometry using Annexin V and propidium iodide staining (Supplementary Fig. 1 and data not shown).
Figure 1. SDS-PAGE electrophoregram of GST-bound proteins (a), PSMA3-bound proteins (b) and cytosol/nuclear extracts (c). “M” lanes: protein molecular mass marker (in kilodaltons).
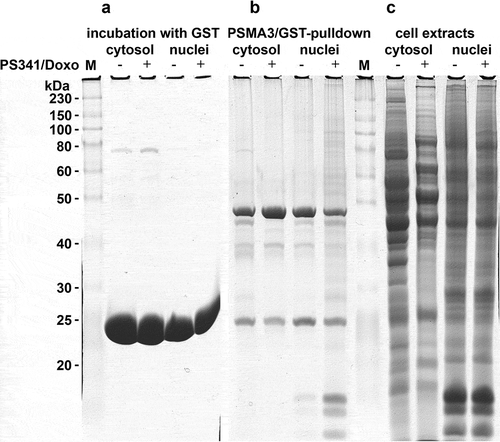
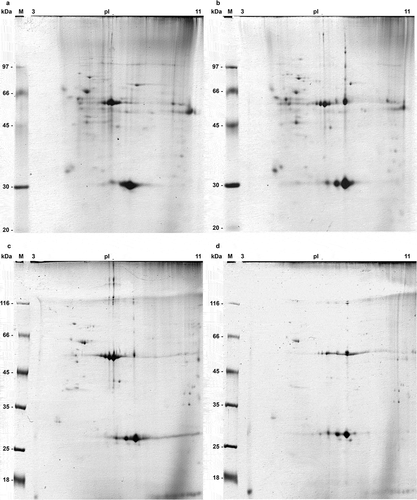
To identify the PSMA3 interacting proteins we employed GST-PSMA3 pull-down technique (). GST beads were used as negative control (). The GST-PSMA3 bound proteins from cytoplasm and the nucleus non-treated and treated with BD were analyzed by 2D electrophoresis (), respectively). Some pKa shifts for certain proteins were also observed, indicating possible changes in post-translational modifications of these proteins.
Figure 2. 2D electrophoregrams of PSMA3-bound proteins from both cytosol (a, b) and nuclear (c, d) extracts of RPMI8226 multiple myeloma cells (control ones (a, c) and after bortezomib/doxorubicin combined treatment (b, d)). Isoelectric focusing was performed in 11 cm IPG strips pH 3–11NL (GE), second dimension was carried out in Hoefer Ruby 600 electrophoresis unit (GE). Gels were stained with Coomassie G250. “M” lane: protein molecular mass marker (in kilodaltons).
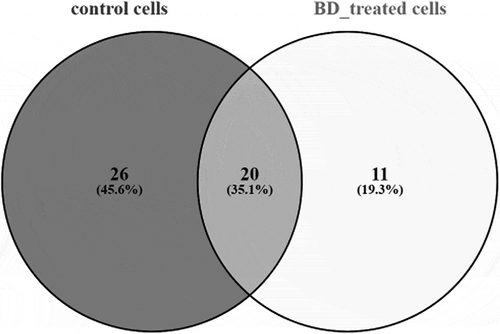
We have identified 46 PSMA3-bound proteins from control RPMI8226 cells and 31 – from cells after combined treatment. 20 proteins were common for both groups, 26 – unique for control cells () and 11 were detected in BD-treated cells () only (). The last group is represented by several cytoskeleton proteins (myosin heavy chain 9, beta 4B tubulin, 47 kDa actin-binding protein), hnRNPs (A1, H2, H3), hemoglobin subunit alpha, histone H2B, coiled-coil domain-containing protein 27, ELAV-like protein 1 and tumor suppressor candidate gene 1 protein.
Table 1. Unique proteins from control RPMI8226 cells interacting with PSMA3.
Table 2. Unique proteins from bortezomib/doxorubicin-treated RPMI8226 cells interacting with PSMA3.
It was possible that several PSMA3-interacting proteins may be degraded both by ubiquitin-independent (via the 20S proteasome) and ubiquitin-dependent (via the 19S sub-complex) pathways. To test such possibility we performed the binding assay of ubiquitinylated proteins with the recombinant HDAC6 ubiquitin interacting domain (HUID) fused to GST. Interestingly, we found five proteins (LSP1, HBA1, HIST1H2BL, HNRNPA1, and ELAVL1) from the validated set of PSMA3 interactors to be associated with HUID suggesting that these proteins may be ubiquitinylated (Supplementary Table 4). Future studies should determine whether ubiquitination affects these interactions and whether these proteins can also be degraded via interactions with subunits of the 19S sub-complex of 26S proteasome. Irrespective of ubiquitinylation, six other proteins (TUSC1, HNRNPH3, HNRNPH2, CCDC27, TUBB4B, and MYH9) apparently interacted with PSMA3 directly, because they were absent from the HUID interactome.
Identification of potential ubiquitin-independent proteolysis substrates using the bioinformatics approach. To undergo rapid degradation via ubiquitin-independent mechanism the target protein must contain an unstructured region (> 30 amino acids in length) [Citation18–Citation20]. Such regions are referred to as intrinsically disordered regions (IDRs). Proteins with entirely disordered sequences (intrinsically disordered proteins, IDPs) do also exist [Citation21]. Bearing this idea in mind, we have evaluated the degree of intrinsic disorder of the identified PSMA3-interacting proteins using DynaMine (http://dynamine.ibsquare.be) [Citation22,Citation23], GlobProt (http://globplot.embl.de) [Citation24] and CSpritz (http://protein.bio.unipd.it/cspritz/) [Citation25] servers. The analysis revealed that half or the proteins that interact with PSMA3 intrinsically carry sufficiently long disordered fragments and thereby can potentially be considered as substrates for ubiquitin-independent proteasomal degradation. The full list of the identified proteins (with IDR-containing members marked) is shown in Supplementary Table 1.
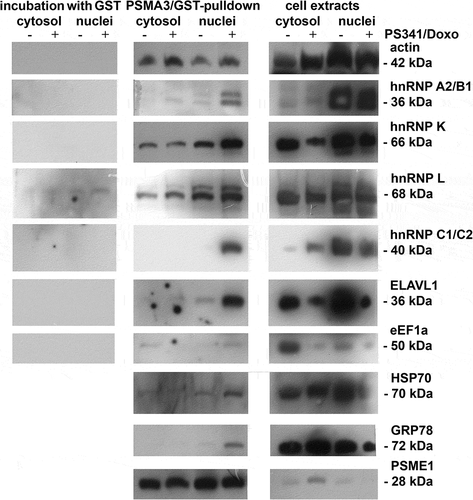
Western blot analysis of PSMA3-interacting proteins. To validate the identities of the PSMA3-interacting proteins we performed Western blotting using the same samples () showed in (1D SDS-PAGE). To confirm the in vitro interactions of cellular proteins with GST-PSMA3 we performed LC-MALDI mass-spectrometry on the proteins associated with purified proteasome samples from RPMI8226 MM cells (274 and 246 proteins were identified, respectively; see Supplementary tables 2 and 3) followed by Western blotting ().
Figure 5. SDS-PAGE electrophoregram (a) and Western blotting against specific antibodies to proteins identified by mass-spectrometry (b) of proteasomes and proteasomal interactome from both control and BD-treated RPMI8226 cells. “M” lane: protein molecular mass marker (in kilodaltons).
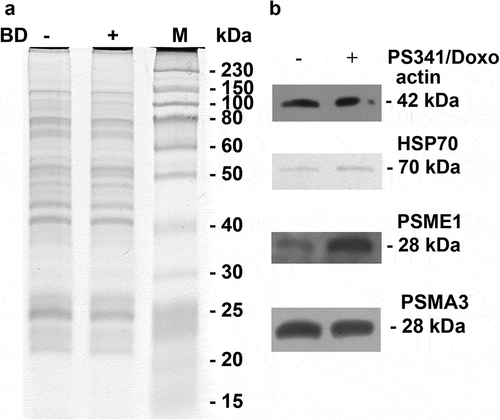
Heterogeneous nuclear ribonucleoproteins belong to a large family of RNA-binding proteins. They play role in pre-mRNA processing and are important in mRNA export, localisation, translation, and stability [Citation26]. Importantly, changes in the expression and distribution patterns of these proteins differ from each other. Whereas HNRNPK and HNRNPL showed similar distribution in all samples, the level of HNRNPK was attenuated in cell extracts after the treatment with BD (). This result may be explained by the fact that HNRNPK participates in the TP53 response to DNA damage and is susceptible to DNA damage response signaling [Citation27]. On the other hand, HNRNP A2/B1 and C1/C2 were found mostly in the nuclear extracts. Interestingly, these proteins were able to interact with PSMA3 only after BD treatment, suggesting that they may undergo post-translational modifications to gain this property.
Intriguingly, we also detected changes in the abundance of the ELAVL1 protein (also known as HuR). This is another RNA binding protein that recognizes AU-rich sequences located in the 3ʹ-UTRs of target genes and protects the respective mRNAs from degradation [Citation28–Citation30]. ELAVL1 also plays role in regulation of IL-3 expression [Citation31]. Abundance of ELAVL1 decreased in both cytoplasm and nucleus after the combined treatment (, column 3). This was concomitant with the increased level of PSMA3 in the nucleus (, column 2).
Similar distribution patterns were observed for HSPA8 (HSP70), and HSPA5 (GRP78) proteins, which are involved in the correct folding of proteins and degradation of misfolded proteins. These proteins also bound PSMA3 only in the nuclear fraction of drug-treated cells (). Intriguingly, HSP70 was already reported to interact with the 19S regulatory particle [Citation32] and can be degraded by the 20S proteasome [Citation33].
Thus, we have identified two populations of proteins that bind the alpha-type PSMA3 subunit of the 20S complex either under normal conditions or after bortezomib/doxorubicin treatment. Most of these proteins contain IDRs, which makes them susceptible to protease-mediated regulation. Therefore, it is likely that these PSMA3-interacting proteins may undergo ubiquitin-independent degradation via the 20S core proteasome complex. This catalog of PSMA3 interactome may be a valuable tool for prediction of the protein stability for specific candidates upon genotoxic stress in MM cancer cells.
Discussion
In the present work, we have identified a number of proteins bound to the PSMA3 subunit of the 20S proteasome core after combined bortezomib/doxorubicin (BD) treatment of human RPMI8226 multiple myeloma cells. The bioinformatics analysis revealed about 60% of the aforementioned proteins contain long-stretched intrinsically disordered regions that were shown previously to be the characteristic feature of substrates for ubiquitin-independent proteasomal degradation [Citation34]. We specifically focused on the proteins that interact with the proteasomal outer core subunit PSMA3 because a number of reports suggested that this particular subunit is the linchpin for various proteins to undergo ubiquitin-independent degradation in the proteasome, including several important oncogenes and tumor suppressor proteins [Citation18,Citation35,Citation36]. Alterations in the degradation dynamics of these proteins may have tumorigenic consequences and these changes may be important for understanding the molecular mechanisms in cancer cells. Indeed, we were able to identify several important interactors with GST-PSMA3 (, ).
Moreover, our bioinformatics analysis suggested that many identified PSMA3-interacting proteins contain intrinsically disordered regions (IDRs), which makes such proteins susceptible for proteolytic degradation (including the proteasome-dependent one). Among the identified IDR-containing proteins is the group of heterogeneous nuclear RNPs that deserves special attention. The proteins of this group not only play role in RNA processing, but also function as transcription factors [Citation37]. For example, HNRNPA1 was reported to take part in the regulation of the cell cycle and proliferation for oral squamous cell carcinoma [Citation38]. HNRNPK was also shown to alter the malignant potential of cancer cells and its aberrant cytoplasmic localization was associated with a negative prognosis for tumor development and an aggressive cell phenotype [Citation39]. Thus, a large number of hnRNPs associate with proteasomes both in the nucleus and cytoplasm of MM cells. Importantly, we validated the identities of PSMA3-interacting proteins revealed by mass-spectrometry by western blotting (). We were able to confirm that most of hnRNPs do bind PSMA3 both as a recombinant protein and as a part of the biochemically isolated proteasome (, and data not shown). Collectively, these results suggest that cycling of hnRNPs may be important for RNA metabolism of MM cells and therefore the search for modulators of hnRNPs stability on the protein level could be a novel therapeutic approach.
In addition, the interaction between the RNA-binding protein HuR/ELAVL1 and PSMA3 appears to be functionally significant. Importantly, similar to HuR, proteasomes are also capable of recognizing AU-rich sequences [Citation2,Citation40]. Furthermore, PSMA3 was shown to exhibit RNAse activity on certain substrates [Citation40,Citation41]. It is tempting to speculate that the interaction between PSMA3 and ELAVL1 is mediated by RNA and is important for RNA metabolism in general.
The recently described potential tumor suppressor TUSC1 was also found in the pool of proteasome-interacting proteins (). This protein functions as a tumor suppressor in a number of human malignancies (stomach cancer, lung cancer and hepatocellular carcinoma) [Citation42–Citation44]. Interestingly, according to the GeneCards (www.genecards.org) data analysis, TUSC1 can also interact with ELAVL1. Noteworthy, TUSC1 was detected as one of the PSMA3-interacting proteins by mass-spectrometry only in the nuclear samples from cells treated with BD (). This result may indicate that TUSC1 is involved in the drug treatment response of MM cells.
The panel of proteins described in this report looks very interesting as most of these proteins contain IDRs in their sequences. Therefore, these PSMA3-interacting proteins may be substrates for ubiquitin-independent proteolysis. However, it should be noted that among those eleven proteins that interact with PSMA3, five of them were also found in the ubiquitome of MM cells because they interacted with the ubiquitin-interacting domain of HDAC6 (Supplementary Table 4). Our results suggest that these proteins may undergo degradation via both ubiquitin-independent and ubiquitin-dependent mechanisms. However, the other six proteins that interact with PSMA3 upon apoptosis, are likely represent novel targets for 20S proteasomes. It is important to note that only a dozen of such substrates are known to date. Interestingly, a large portion of nuclear proteins associate with PSMA3 only after the combined treatment with bortezomib and doxorubicin (). Several of the identified proteins are known as putative biomarkers for a number of cancers (for example, GRP78, PDIA6) [Citation45,Citation46], epithelial-mesenchymal transition markers (VIM) and are considered as perspective therapeutic targets (HSP70, PHB) [Citation47,Citation48]. Thus, it will be important to determine whether the forced accumulation of these proteins can cause proteotoxic stress and sensitize MM cells to genotoxic therapy.
Materials and methods
Cell cultures. Human multiple myeloma RPMI8226 cells [Citation49] obtained from the Russian Collection of Cell Cultures (Institute of Cytology of RAS) were cultured at 37°C in RPMI 1640 medium containing 10% fetal calf serum in the presence of 0.004% gentamycin. Treatment of cells with the combination of doxorubycin (2 uM) and bortezomib (10 nM) was conducted for 16 hours.
Isolation of proteasomes. Cells were pelleted by centrifugation (1000 min−1, 10 min, 4°C) and stored at −70°C. Proteasomes were isolated from cell cytosol by centrifugation in sucrose gradient (15–30%) followed by anion-exchange chromatography on DEАЕ cellulose [19]. The peptidase activities of proteasomes were measured fluorometrically using the following fluorogenic peptides as substrates: Benzyloxycarbonyl-L-leucyl-L-leucyl-glutamyl-L-7-amino-4-methylcoumarin – Bz-LLE-AMC (caspase-like activity), t-butoxycarbonyl-L-leucyl-L-arginyl-L-arginyl-methylcoumarylamid – Boc-LRR-AMC (trypsin-like) and N-Succinyl-L-leucyl-L-leucyl-L-valyl-L-tyrosine-methylcoumarylamid – Suc-LLVY-AMC (chymotrypsin-like) (Biomol, UK). All three peptidase activities decreased after bortezomib treatment of RPMI8226 cells: trypsin-like – 1.5-fold, chymotrypsin-like – 3.5-fold and caspase-like – 2.6-fold.
Purification of the fusion proteins GST-PSMA3, GST-HUID and GST pull-down. The GST-PSMA3 and GST-HUID expression vectors were obtained by insertion of the human PSMA3 and polyubiquitin interacting domain of the human HDAC6 protein (HUID) cDNAs, respectively, into the pGEX-5X vector [Citation50]. The recombinant proteins were expressed in the BL21 E. coli strain by adding 0.2 mM IPTG for 3 hours at 37°C. The bacterial extracts were clarified by centrifugation and subsequently incubated with glutathione-sepharose 4B beads for one hour (GE Healthcare, USA). To purify the proteins associated with GST-PSMA3 and GST-HUID both cytosol and nuclear extracts were prepared from RPMI8226 multiple myeloma cells and incubated with affinity resins. GST alone was used as control.
Two-dimensional electrophoresis of proteasome-associated proteins. Isoelectric focusing was performed in IPG strips (11 cm long, pH 3–11 NL) in IPG Phor 3 IEF system (GE Healthcare) followed by SDS-PAGE separation according to Laemmli [Citation51] with some modifications. The proteins were visualized by Coomassie Brilliant blue G250 staining.
Western-blotting. After separation in SDS-PAGE (AA: 10%, AA/BisAA ratio: 36:1), the proteins were transferred onto PVDF membranes following overnight incubation with specific primary antibodies. The following antibodies were used: HSP70 – mAb ADI-SPA-810 (Enzo; http://www.enzolifesciences.com/ADI-SPA-810/hsp70-hsp72-monoclonal-antibody-c92f3a-5/), PSMA3 – Mouse mAb BML-PW8110 (Enzo; http://www.enzolifesciences.com/BML-PW8110/proteasome-20s-alpha7-subunit-monoclonal-antibody-mcp72/), PSME1 – Rabbit BML-PW8185 (Enzo; http://www.enzolifesciences.com/BML-PW8185/proteasome-activator-11s-alpha-subunit-polyclonal-antibody/), ELAVL1/HuR – Rabbit mAb #12,582 (Cell signaling; https://www.cellsignal.com/products/primary-antibodies/elavl1-hur-d9w7e-rabbit-mab/12582), β-Actin – Mouse mAb #3700 (Cell signaling; https://www.cellsignal.com/products/primary-antibodies/b-actin-8h10d10-mouse-mab/3700), eEF1A – Rabbit mAb #3586 (Cell signaling; https://www.cellsignal.com/products/primary-antibodies/eef1a-d10a5-rabbit-mab/3586), hnRNP A2/B1 – Mouse mAb #9304 (Cell signaling; https://www.cellsignal.com/products/primary-antibodies/hnrnp-a2-b1-2a2-mouse-mab/9304), hnRNP K – Rabbit ab18195 (Abcam; http://www.abcam.com/hnrnp-k-antibody-ab18195.html), hnRNP C1/C2 – Rabbit ab97541 (Abcam; http://www.abcam.com/hnrnp-c1-c2-antibody-ab97541.html), hnRNP L – Rabbit ab32680 (Abcam; http://www.abcam.com/hnrnp-l-antibody-ab32680.html), GRP78 – Rabbit ab188878 (Abcam; http://www.abcam.com/grp78-bip-antibody-ab188878.html).
Mass-spectrometry. Trypsin digested (Trypsin Gold, Promega, USA) proteins after preliminary desalting were analyzed by AB Sciex 5800 MALDI TOF/TOF mass-spectrometer (AB Sciex). Fragment ion MS/MS spectra were searched by MASCOT search tool against the UniProtKB/Swiss-Prot protein database using appropriate parameters.
Bioinformatics. The evaluation of the protein degree of linear disorder was performed using four independent sources. Protein backbone dynamics prediction servers – DynaMine (http://dynamine.ibsquare.be) [Citation22,Citation23], GlobProt (http://globplot.embl.de) [Citation24] and CSpritz (http://protein.bio.unipd.it/cspritz/) [Citation25] that exploit different algorithms to predict the degree of disorder – were used together with the IDEAL database of experimentally verified intrinsically disordered proteins [Citation52]. The cutoff value of a single longest sequence, predicted to be disordered was taken as 30 amino acids [Citation53], however some data from different sources were not in full agreement and therefore required manual curation based on the data from literature.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Supplemental Material
Download Zip (540.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary materials
Supplementary data for this article can be accessed here.
Additional information
Funding
References
- Mittenberg AG, Moiseeva TN, Barlev NA. Role of proteasomes in transcription and their regulation by covalent modifications. Front Biosci. 2008;13:7184–7192. PMID: 18508726.
- Mittenberg AG. Non-canonical activities of proteasomes. Tsitologiia. 2014 in Russian;56:331–339. PMID: 25696972.
- Tsimokha AS, Kulichkova VA, Karpova EV, et al. DNA damage modulates interactions between microRNAs and the 26S proteasome. Oncotarget. 2014;5:3555–3567. PMID: 25004448.
- Moiseeva TN, Bottrill A, Melino G, et al. DNA damage-induced ubiquitylation of proteasome controls its proteolytic activity. Oncotarget. 2013;4:1338–1348. PMID: 23907514.
- Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J Mol Biol. 2009;386:814–827. PMID: 19162040.
- Murakami Y, Matsufuji S, Kameji T, et al. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. PMID: 1334232.
- Tsvetkov P, Reuven N, Shaul Y. Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ. 2010;17:103–108. PMID: 19557012.
- Malatesta M, Peschiaroli A, Memmi EM, et al. The Cul4A-DDB1 E3 ubiquitin ligase complex represses p73 transcriptional activity. Oncogene. 2013;32:4721–4726. PMID: 23085759.
- Barlev NA, Sayan BS, Candi E, et al. The microRNA and p53 families join forces against cancer. Cell Death Differ. 2010;17:373–375. PMID: 20062068.
- Cayrol C, Ducommun B. Interaction with cyclin-dependent kinases and PCNA modulates proteasome-dependent degradation of p21. Oncogene. 1998;17:2437–2444. PMID: 9824154.
- Touitou R, Richardson J, Bose S, et al. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 α-subunit of the 20S proteasome. EMBO J. 2001;20:2367–2375. PMID: 11350925.
- Sdek P, Ying H, Chang DLF, et al. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell. 2005;20:699–708. PMID: 16337594.
- Bae MH, Jeong CH, Kim SH, et al. Regulation of Egr-1 by association with the proteasome component C8. BBA. 2002;1592:163–167. PMID: 12379479.
- Konstantinova IM, Tsimokha AS, Mittenberg AG. Role of proteasomes in cellular regulation. Int Rev Cell Mol Biol. 2008;267:59–124. PMID: 18544497.
- Moiseeva TN, Mittenberg AG, Barlev NA. Proteasomes and their role in transcriptional regulation. Tsitologiia. 2010;52:195–203. in Russian. PMID: 20429296.
- Hideshima T, Richardson P, Chauhan D, et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human. Multiple Myeloma Cells Cancer Res. 2001;61:3071–3076. PMID: 11306489.
- Obeng EA, Carlson LM, Gutman DM, et al. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–4916. PMID: 16507771.
- Hoyt MA, Coffino P. 5. ubiquitin-independent mechanisms of substrate recognition and degradation by the proteasome. Protein Degradation: the Ubiquitin-Proteasome System and Disease. 2007;4:107–121.
- Hwang J, Winkler L, Kalejta RF. Ubiquitin-independent proteasomal degradation during oncogenic viral infections. Biochim Biophys Acta. 2011;1816:147–157. PMID: 21664948.
- Pickering AM, Davies KJ. Degradation of damaged proteins: the main function of the 20S proteasome. Prog Mol Biol Transl Sci. 2012;109:227–248. PMID: 22727423.
- Van Der Lee R, Buljan M, Lang B, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. PMID: 24773235.
- Cilia E, Pancsa R, Tompa P, et al. From protein sequence to dynamics and disorder with dynamine. Nat Commun. 2013;4:2741. PMID: 24225580.
- Cilia E, Pancsa R, Tompa P, et al. The dynamine webserver: predicting protein dynamics from sequence. Nucl Acids Res. 2014;42:W264–W270. PMID: 24728994.
- Linding R, Russell RB, Neduva V, et al. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. PMID: 12824398.
- Walsh I, Martin AJ, Di Domenico T, et al. CSpritz: accurate prediction of protein disorder segments with annotation for homology, secondary structure and linear motifs. Nucleic Acids Res. 2011;39:W190–W196. PMID: 21646342.
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. PMID: 11994740.
- Moumen A, Masterson P, O’Connor MJ, et al. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. PMID: 16360036.
- Ma WJ, Cheng S, Campbell C, et al. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. PMID: 8626503.
- Tran H, Maurer F, Nagamine Y. Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol Cell Biol. 2003;23:7177–7188. PMID: 14517288.
- Wang H, Zeng F, Liu Q, et al. The structure of the ARE-binding domains of hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Crystallographica Section D: Biological Crystallography. 2013;69:373–380. PMID: 23519412.
- González-Feliciano JA, Hernández-Pérez M, Estrella LA, et al. The role of HuR in the post-transcriptional regulation of interleukin-3 in T cells. PLoS One. 2014;9(3):e92457. PMID: 24658545.
- Grune T, Catalgol B, Licht A, et al. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radic Biol Med. 2011;51:1355–1364. PMID: 21767633.
- Morozov AV, Astakhova TM, Garbuz DG, et al. Interplay between recombinant Hsp70 and proteasomes: proteasome activity modulation and ubiquitin-independent cleavage of Hsp70. Cell Stress Chaperones. 2017;22:687–697. PMID: 28447215.
- Erales J, Coffino P. Ubiquitin-independent proteasomal degradation. PMID: 23684952 Biochim Biophys Acta. 2014;18431:216–221.
- Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta - Reviews on Cancer. 2008;1786:153–177. PMID: 18558098.
- Fedorova OA, Moiseeva TN, Nikiforov AA, et al. Proteomic analysis of the 20S proteasome (PSMA3)-interacting proteins reveals a functional link between the proteasome and mRNA metabolism. Biochem Biophys Res Commun. 2011;416:258–265. PMID: 22079093.
- Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1’s multifunctional regulatory roles. RNA. 2010;16:1449–1462. PMID: 20584894.
- Yu C, Guo J, Liu Y, et al. Oral squamous cancer cell exploits hnRNP A1 to regulate cell cycle and proliferation. J Cell Physiol. 2015;230:2252–2261. PMID: 25752295.
- Barboro P, Ferrari N, Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 2014;352:152–159. PMID: 25016060.
- Va K, Oa F, As T, et al. 26S proteasome exhibits endoribonuclease activity controlled by extra-cellular stimuli. Cell Cycle. 2010;9:840–849. PMID: 20139718.
- Mittenberg AG, Moiseeva TN, Kuzyk VO, et al. Regulation of endoribonuclease activity of alpha-type proteasome subunits in proerythroleukemia K562 upon hemin-induced differentiation. Protein J. 2016;35:17–23. PMID: 26661102.
- Shan Z, A S, Bodaghi S, et al. TUSC1, a putative tumor suppressor gene, reduces tumor cell growth in vitro and tumor growth in vivo. PLoS One. 2013;8:e66114. PMID: 23776618.
- Kanda M, Shimizu D, Nomoto S, et al. Clinical significance of expression and epigenetic profiling of TUSC1 in gastric cancer. J Surg Oncol. 2014;110:136–144. PMID: 24700496.
- Shimizu D, Kanda M, Nomoto S, et al. Identification of intragenic methylation in the TUSC1 gene as a novel prognostic marker of hepatocellular carcinoma. Oncol Rep. 2014;31:1305–1313. PMID: 24366000.
- Guan M, Chen X, Ma Y, et al. MDA-9 and GRP78 as potential diagnostic biomarkers for early detection of melanoma metastasis. Tumour Biol. 2015;36:2973–2982. PMID: 25480418.
- Ramos FS, Serino LT, Carvalho CM, et al. PDIA3 and PDIA6 gene expression as an aggressiveness marker in primary ductal breast cancer. Genet Mol Res. 2015;14:6960–6967. PMID: 26125904.
- Shevtsov M, Huile G, Multhoff G. Membrane heat shock protein 70: a theranostic target for cancer therapy. Philos Trans R Soc Lond B Biol Sci. 2018;373(1738):20160526. pii.
- Guo S, Zou J, Wang G. Advances in the proteomic discovery of novel therapeutic targets in cancer. Drug Des Devel Ther. 2013;7:1259–1271. PMID: 24187485.
- Matsuoka Y, Moore GE, Yagi Y, et al. Production of free light chains of immunoglobulin by a hematopoietic cell line derived from a patient with multiple myeloma. Proc Soc Exp Biol Med. 1967;125:1246–1250. PMID: 6042436.
- Mittenberg AG, Moiseeva TN, Barlev NA. Method of identifying the short-living regulatory proteins in human proteome involved in the specific modulation of cytotoxic effect. Inventions. Useful Models. Official Bulletin of Federal Service for Intellectual Property (Russia). 2016;19(2590700):1–12.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227: 680–685. PMID: 5432063.
- Fukuchi S, Amemiya T, Sakamoto S, et al. IDEAL in illustrates interaction networks composed of intrinsically disordered proteins and their binding partners. Nucleic Acids Res. 2014;42:D320–D325. PMID: 24178034.
- Ben-Nissan G, Sharon M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules. 2014;4:862–884. PMID: 25250704.