ABSTRACT
Protein phosphorylation regulates a variety of cellular signaling pathways and fundamental mechanisms in cells. In this paper, we demonstrate that the mRNA decay factor Roquin2 is phosphorylated at tyrosine residue in position 691 in vivo. This phosphorylation disrupts the interaction with KLHL6, the E3 ligase for Roquin2. Furthermore, we establish that the tyrosine phosphatase PTPN14 specifically interacts with Roquin2 through its phosphatase domain and dephosphorylates Roquin2 tyrosine 691. Overexpression of PTPN14 promotes Roquin2 degradation in a KLHL6-dependant manner by promoting interaction with KLHL6. Collectively, our findings reveal that PTPN14 negatively regulates the protein stability of Roquin2, thereby adding a new layer of regulation to the KLHL6-Roquin2 axis.
Introduction
Protein post-translational modifications (PTMs) are pivotal in regulating enzyme activity and signaling transduction in cells. Amongst the PTMs, protein phosphorylation and ubiquitylation are considered the most abundant in eukaryotes [Citation1–Citation3]. Phosphorylation occurs on serine, threonine, and tyrosines residues through phosphoester bond formation, and it is dynamically regulated by the activity of kinases and phosphatases [Citation3]. Protein ubiquitylation is catalyzed in a stepwise enzymatic cascade through the coordinated activity of an E1 ubiquitin activating-enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin-ligating enzyme [Citation4,Citation5]. These enzymes act together in a precise and specific manner to target proteins for proteasomal degradation [Citation6].
Kelch-like protein 6 (KLHL6) is a member of the bric-a-brac/tramtrack/broad-complex (BTB) domain family of proteins, which assembles a functional CULLIN3-E3 ubiquitin ligase [Citation7]. The KLHL6 gene displays a high frequency of inactivating mutations in diffuse large B-cell lymphoma (DLBCL) [Citation7–Citation11]. Loss of KLHL6 catalytic activity results in the stabilization of the mRNA decay factor Roquin2 and the activation of NF-κB pathway in DLBCL cells [Citation7]. Roquin2 belongs to the Roquin family of proteins that consists of Roquin1 (RC3H1) and Roquin2 (RC3H1) [Citation12] and promotes mRNA decay of transcripts containing a constitutive-decay element (CDE) in their 3ʹ UTR [Citation13–Citation15]. Our previous study revealed that KLHL6 recognizes a tyrosine in position 691 on Roquin2, and mutation into phenylalanine [Roquin2(Y691F)] stabilizes the protein and promotes DLBCL cell proliferation [Citation7]. Whether or not phosphorylation regulates the KLHL6-Roquin2 interaction and protein stability remains an open question.
Protein tyrosine phosphatase non-receptor type 14 (PTPN14) has an N-terminal FERM and a C-terminal phosphatase domain, and it plays a critical role in cellular proliferation and growth, cell-cell adhesion, and cell migration [Citation16–Citation18]. In addition, recent studies have shown that PTPN14 is mutated in multiple types of cancers including colorectal, breast, ovarian, and liver cancers [Citation19–Citation22]. Mechanistically, PTPN14 inhibits the oncogenic function of the yes-associated protein 1(YAP1), a protein involved in the hippo signaling pathway [Citation22,Citation23] and suppresses both pancreatic cancer progression and metastasis in breast cancer [Citation24,Citation25]. Only three substrates (YAP, B-catenin, and p130Cas) have been identified so far, and the other biological functions of PTPN14 in different genetic and cellular contexts remain to be determined [Citation17,Citation19,Citation23].
We demonstrate that Roquin2 is phosphorylated at tyrosine residue 691 in cells, and this phosphorylation disrupts the interaction between Roquin2 and KLHL6. We present evidence that PTPN14 de-phosphorylates Roquin2 on the tyrosine residue 691. Overexpression of the wild-type PTPN14 in B-cell lymphomas decreases the protein abundance of Roquin2 as compared to an inactive mutant of PTPN14. These findings reveal that PTPN14 is a novel regulator of Roquin2 stability and suggest that phosphorylation modulates the degradation of Roquin2.
Results
Phosphorylation of tyrosine 691 in Roquin2 inhibits interaction with KLHL6
Our previous study showed that the integrity of tyrosine 691 (Y691) residue in Roquin2 is critical for KLHL6 interaction, as the mutagenesis of this residue to alanine and phenylalanine impacts the KLHL6-Roquin2 interaction [Citation7]. Since the tyrosine residue can be modified post-translationally by phosphorylation [Citation3], we decided to validate it. To this purpose, we generated an antibody recognizing phospho-tyrosine 691 of Roquin2 and tested HEK293T cells transfected with FLAG-STREP-tagged wild-type (WT) or single tyrosine point mutant Roquin2 (Y691F), a non-phosphorylatable form of tyrosine. To increase the level of cellular tyrosine phosphorylation, we treated cells with pervanadate [Citation26,Citation27], (). Pretreatment of cells with pervanadate induced a strong phosphorylation of proteins in tyrosine, which was detected by the analysis of the whole cell lysates with a phospho-tyrosine antibody ()). Following the pervanadate treatment, our phospho-antibody recognized Roquin2 (WT) but not the Roquin2 (Y691F) mutant ()). To further evaluate the phospho-specificity of our antibody, we dephosphorylated Roquin2 (WT) with λ-phosphatase (). This treatment abrogated the ability of our antibody to recognize Roquin2 (WT) when isolated from pervanadate treated cells, suggesting that it indeed recognizes a phosphorylated moiety in position 691. Notably, the antibody could not detect the phosphorylation of tyrosine 691 in the steady state. This could imply that either the basal level of Roquin2 phosphorylation is very low or our antibody is not sensitive enough to detect the basal phosphorylation.
Figure 1. Tyrosine 691 in Roquin2 is phosphorylated in vivo.
(a) HEK293T cells were transfected with cDNAs encoding FLAG-STREP-tagged Roquin2 (WT), Roquin2 (Y691F) or empty vector (EV). The transfected cells were treated with 100µM of pervanadate for 15 min and whole cell lysates were used for immunoblot analysis. PerV, Pervanadate. (b) Exogenous proteins were affinity-purified (AP) from cell extracts of (a) with an anti-streptavidin resin in a denatured condition. The purified complexes were, then, treated with lamda (λ)-phosphotase and probed with antibodies to the indicated proteins. PerV, Pervanadate. (c) HEK293T cells stably expressing HA-tagged KLHL6 were transfected with cDNAs encoding FLAG-tagged Roquin2 (WT) or empty vector (EV). Exogenous proteins were immunopurified from cell extracts with an anti-FLAG resin and immunocomplexes were probed with antibodies to the indicated proteins. Bottom panel shows whole cell lysates (WCL). PerV, Pervanadate. (d) Schematic representation of the sequence of the biotinylated unphosphosphorylated-Roquin2 peptides or phosphorylated-Roquin2 peptides. (e) Streptavidin pull-down assay using the indicated amount of biotinylated Roquin2 peptides incubated with FLAG-tagged in-vitro translated KLHL6 protein. Immunoblot analysis for the indicated proteins was performed using KLHL6 antibody. Affinity Purification, AP. STREP, Streptavidin.
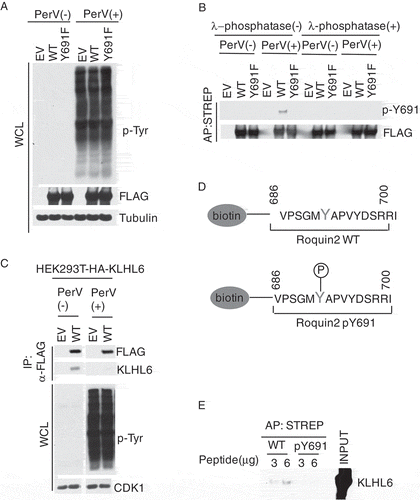
Many PTMs function to regulate the protein-protein interaction[Citation28]. Given that tyrosine 691 in Roquin2 is critical for interaction with KLHL6 [Citation7], we decided to assess the impact of phosphorylation on this interaction. Since we have previously found that tyrosine 691 is phosphorylated upon pervanadate treatment ()), we examined the interaction between Roquin2 and KLHL6 in the same condition. Interestingly, the pervanadate treatment disrupted the binding between Roquin2 and KLHL6, suggesting that tyrosine phosphorylation negatively regulates the interaction between the two proteins ()). To directly evaluate whether tyrosine phosphorylation inhibits the KLHL6-Roquin2 interaction, we synthetized peptides containing unphospho- and phospho-tyrosine 691 ()). Using an in vitro binding assay, we demonstrated that phosphorylation of tyrosine 691 impaired the ability of Roquin2 to associate with KLHL6 ()), while the unphosphorylated-peptide efficiently pulled down KLHL6.
In conclusion, our data suggests that the tyrosine in position 691 of Roquin2 is phosphorylated in cells. Moreover, phosphorylation at tyrosine 691 in Roquin2 negatively regulates the KLHL6-Roquin2 interaction.
PTPN14 specifically interacts with Roquin2
Since Roquin2, but not its paralog Roquin1, specifically interacts with KLHL6, we hypothesized that tyrosine 691 could be modified by a kinase or phosphatase specific to Roquin2. In order to identify Roquin2-specific interactors, we compared the protein interactome of Roquin1 to that of Roquin2 () and Supplementary Table 1). FLAG-Roquin1 or FLAG-Roquin2 complexes were immunopurified, and the tryptic digestion of each protein eluate was analyzed using mass spectrometry. Our proteomic analysis validated the known Roquin1 and Roquin2 interactors as the deadenylation factors (CNOT1,2,3,7,10,11) and the ribosomal proteins (RPL3-38/RPS2-29) () and [Citation13,Citation29–Citation31]). Additionally, we identified PTPN14 as a specific binding partner of Roquin2 ()).
Figure 2. PTPN14 specifically interacts with Roquin2.
(a) Biochemical purification of Roquin1 or Roquin2 protein complexes. HEK293T cells were transfected with cDNAs encoding FLAG-STREP Roquin1 (WT) or FLAG-STREP Roquin2 (WT). Proteins were immunoprecipitated (IP) with an anti-FLAG resin (α-FLAG), eluted with a FLAG peptide. 1% of samples were resolved by SDS-PAGE. The gel was stained with silver staining for protein visualization. Asterisks indicate the bait. (b) Scatter plot of identified proteins by mass spectrometry analysis of Roquin1 (WT) and Roquin2 (WT). Normalized Spectral Abundance Factors (NSAFs) were calculated for each detected protein and plotted on a log scale. X-axis represents NSAF scores distribution of all proteins detected from Roquin2 protein complexes while Y-axis represents NSAF scores distribution of all proteins detected from Roquin1 protein complexes. Red dots represent NSAF scores for the baits such as Roquin1 and Roquin2. The purple dot represents the NSAF score for PTPN14. The green and black dots represent common interactors between Roquin1 and Roquin2. (c) HEK293T cells were transfected with cDNAs encoding empty vector (EV), FLAG-STREP tagged Roquin1 or FLAG-STREP tagged Roquin2. Exogenous proteins were immunopurified from cell extracts with an anti-FLAG resin and immunocomplexes were probed with antibodies to the indicated proteins. Bottom panel shows whole cell lysates (WCL).
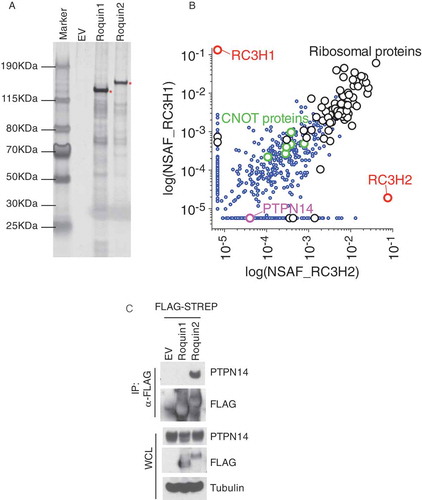
PTPN14 is a non-receptor type of tyrosine phosphatase [Citation16]. In order to validate the proteomic analysis, we expressed and immunoprecipitated FLAG-tagged Roquin1 or Roquin2 from HEK293T cells and confirmed the interaction of endogenous PTPN14 specifically with Roquin2. Notably, Roquin1, although expressed at a higher level, was incapable of binding with PTPN14 ()). In conclusion, we identified PTPN14 as a specific interactor of Roquin2.
PTPN14 binds the C-terminal region of the Roquin2 protein through its phosphatase domain
To determine the regions of Roquin2 that contributed to its interaction with PTPN14, we generated a set of C-terminal deletion mutants in Roquin2 using site-directed mutagenesis. Given that Roquin1 and Roquin2 have a high sequence similarity including the RING, ROQ, and Zinc Finger domains [Citation32] and that PTPN14 specifically interacts with Roquin2, we predicted that the binding region should be confined to the C-terminal region of Roquin2. Consistent with our hypothesis, C-terminal deletions eliminated the ability of Roquin2 to bind to PTPN14 up to the mutant lacking 50 amino acids at the C-Terminal ()). Notably, Roquin1 differs from Roquin2 in its C-Terminal region, which features a coiled-coil domain rather than a hydrophobic-rich region responsible for binding with PTPN14 [Citation32].
Figure 3. PTPN14 binds the C-terminal region of Roquin2 through the phosphatase domain.
(a) Left, schematic representation of Roquin2 mutants and binding to PTPN14. Roquin2 mutants that interact (+) or do not interact (-) with endogenous PTPN14 are shown. Right, immunoblot analysis of FLAG-Roquin2 immunoprecipitation (IP). HEK293T cells were transfected with constructs encoding an empty vector (EV), FLAG-tagged Roquin2 (WT) or FLAG-tagged Roquin2 deletion mutants as indicated. Immunocomplexes were probed with antibodies to the indicated proteins. STREP, Streptavidin. (b) Left, schematic representation of PTPN14 mutants and binding to Roquin2. PTPN14 mutants that interact (+) or do not interact (-) with exogenous Roquin2 are shown. Right, immunoblot analysis of FLAG-Roquin2 immunoprecipitation (IP). HEK293T cells were co-transfected with constructs encoding an empty vector (EV), V5-tagged PTPN14 (WT) or V5-tagged PTPN14 deletion mutants and FLAG-tagged Roquin2 as indicated. Immunocomplexes were probed with antibodies to the indicated proteins.
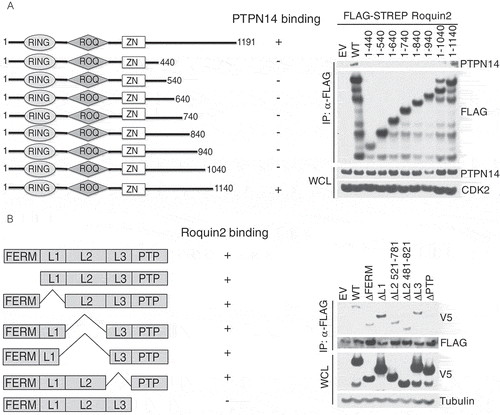
To delineate this interaction on PTPN14, we utilized a panel of PTPN14 deletion mutants. These mutants included deletions of FERM domain, the linker 1 region, the linker 2 region, the linker 3 region, and the phosphatase (PTP) domain [Citation22,Citation33,Citation34]. We co-transfected HEK293T cells with N-terminal V5-tagged PTPN14 mutants with FLAG-tagged Roquin2, and FLAG immunoprecipitates were probed with V5 and FLAG antibodies. FLAG-Roquin2 efficiently co-immunoprecipitated the wild-type and all the deletion mutants of PTPN14 excluding the mutant lacking the PTP domain ()). Thus, we concluded that the phosphatase domain of PTPN14 is required to bind the C-terminus of Roquin2.
PTPN14 enhances interaction between KLHL6 and Roquin2 by directly dephosphorylating Y691
Next, we investigated whether PTPN14 affects the interaction between KLHL6 and Roquin2. To this purpose, we transfected HEK293T cells, stably expressing HA-KLHL6 with FLAG-tagged Roquin2 and V5-tagged PTPN14. Cell lysates were immunoprecipitated with anti-FLAG, and the precipitates were probed with FLAG, KLHL6, or V5 antibodies. As expected, FLAG-Roquin2 co-immunopurified with both V5-tagged PTPN14 and HA-tagged KLHL6 ()). Furthermore, the expression of PTPN14 increased the binding of KLHL6 and Roquin2, suggesting that PTPN14 favors the KLHL6 and Roquin2 interaction.
Figure 4. PTPN14 regulates Roquin2 protein stability in a KLHL6-dependent manner by directly dephosphorylating tyrosine 691.
(a) HEK293T cells stably expressing HA-tagged KLHL6 were co-transfected with cDNAs encoding FLAG-tagged Roquin2 and V5-tagged PTPN14. Exogenous Roquin2 proteins were immunopurified from cell extracts with an anti-FLAG resin and immunocomplexes were probed with antibodies to the indicated proteins. Bottom panel shows whole cell lysates (WCL). HA, hemagglutinin. (b) The indicated FLAG-tagged proteins were immunopurified from HEK293T cell extracts with an anti-FLAG resin. Cells transfected with FLAG-tagged Roquin2 were pre-treated with 100µM of pervanadate for 15 min before collection. Purified proteins on the beads were mixed as indicated for in vitro phosphatase assay. Lamda (λ)-phosphotase treatment was used as a positive control for the assay. Proteins were probed with antibodies to the indicated proteins. Red asterisks indicate the FLAG-tagged proteins immunoblotted with an anti-FLAG antibody. (c) HEK293T cells were co-transfected with cDNAs encoding FLAG-HA-tagged Roquin2 (WT), FLAG-tagged KLHL6, and increasing amounts of V5-tagged PTPN14 in different combinations. Whole cell lysates were probed with antibodies to the indicated proteins. (d) Immunoblot analysis of whole-cell lysates from BJAB cells KLHL6+/+ or KLHL6−/- (clone derived) for the indicated serum starvation time points. (e) Same as in (d) except that BJAB cells stably expressing Roquin2(WT) or Roquin2(Y691F) were used. (f) OCI-LY10 cells were lenti-virally transduced to express doxycycline (DOX) controllable HA-tagged PTPN14 (WT) or PTPN14 (D1079A) mutant. DOX was pre-added for 12 hours and, then, cells were stimulated with anti-IgM (10µg/ml) with indicated time points. Whole cell lysates were probed with antibodies to the indicated proteins. CTRL, control. (g) A schematic model of the complex composed of Roquin2, KLHL6 and PTPN14. PTPN14 binds Roquin2 through its phosphatase (PTP) domain and KLHL6 binds Roquin2 at Y691 residue. PTPN14 regulates Roquin2 and KLHL6 interaction by direct dephosphorylation of Y691 in Roquin2. Phosphorylation, P.
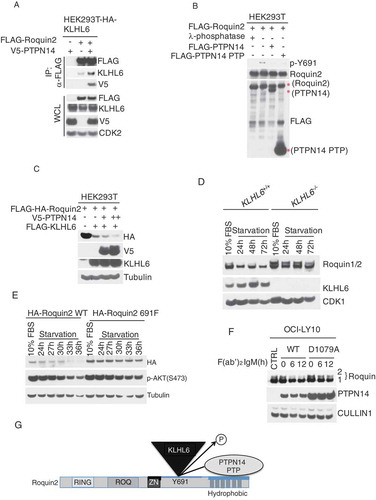
Given that phosphorylation at tyrosine 691 inhibits the recruitment of KLHL6, we hypothesized that PTPN14 dephosphorylates tyrosine 691 to bring KLHL6 and Roquin2 together. To this end, we performed an in vitro phosphatase assay by incubating purified Roquin2, PTPN14 WT, and the PTP domain of PTPN14 ()). PTPN14 WT and its PTP domain dephosphorylated Roquin2 at the tyrosine 691 similar to λ-phosphatase ()), suggesting that PTPN14 directly dephosphorylates Roquin2.
Following, we investigated whether PTPN14 promotes the degradation of Roquin2 via KLHL6. Expression of KLHL6 induced the degradation of Roquin2, this which was enhanced when PTPN14 was co-expressed. This suggests that PTPN14 promotes Roquin2 degradation via KLHL6 ()).
PTPN14 localizes in the cytoplasm upon serum starvation [Citation18]. Therefore, we enquired if serum starvation could change Roquin2 protein levels. Endogenous Roquin2 protein was downregulated upon serum withdrawal, and this effect was not observed in KLHL6−/- B-lymphoma cell lines ()). Correspondingly, the protein levels of the Roquin2 (Y691F) mutant, which does not interact with KLHL6 [Citation7], were minimally affected by serum starvation as compared to Roquin2(WT) ()). Both data suggest that serum starvation induces Roquin2 degradation in a KLHL6-dependent manner.
Furthermore, we analyzed the effect of PTPN14 (WT), or a catalytically inactive mutant PTPN14 (D1079A) [Citation19,Citation35], on the kinetics of Roquin2 levels. We induced Roquin2 degradation by treating DLBCL cells with the fragment affinity-purified antibody F(ab’)2-IgM to activate B-cell receptor (BCR), signaling the pathway as previously shown [Citation7,Citation36]. In cells expressing PTPN14 (WT), Roquin2 was degraded upon BCR stimulation, while this degradation was attenuated in cells expressing PTPN14 (D1079A) ()).
To conclude, our data suggests a model where PTPN14 binds at the hydrophobic-rich region of Roquin2 and functions to promote Roquin2 dephosphorylation and KLHL6-dependent protein degradation ()).
PTPN14 is a putative tumor suppressor in DLBCL
Next, we investigated whether PTPN14 impacts DLBCL cell growth. The expression of PTPN14 (WT) via a doxycycline inducible promoter decreased proliferation of DLBCL cells, while the expression of PTPN14 (D1079A) had no effect (.
Figure 5. PTPN14 is a putative tumor suppressor in DLBCL (A) Cell proliferation of OCI-LY10 cells (from )) that were grown in media containing DOX was monitored by MTS assay and normalized on empty vector at day 1. Error bars represent s.d., n = 3. OD, optical density. (B) Scatter Plot showing the log2 expression of RNA-seq data for KLHL6 and PTPN14 from Cancer Cell Line Encyclopedia (CCLE, https://portals.broadinstitute.org/ccle, n = 1036). The red circles represent DLBCL cell lines. (C) Contingency table to summarize the relationship between PTPN14 and KLHL6 expression (n = 1036, Chi-square). (D) Plot of the log2 expression of RNA-seq data for KLHL6 (left panel) and PTPN14 (right panel) from the CCLE. Cell lines were grouped by tumor type. (E) RNA-seq data from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) for the indicated transcripts (n = 48). (F) A heat map showing the gene expression profile of PTPN14 and KLHL6 in DLBCL tumors from TCGA (n = 48). (G) Kaplan–Meier analysis based on gene expression data for DLBCL tumors (GSE10846) using probe 226282_at is shown (n = 420, Mantel–Cox). Censored subjects are indicated as tick marks.
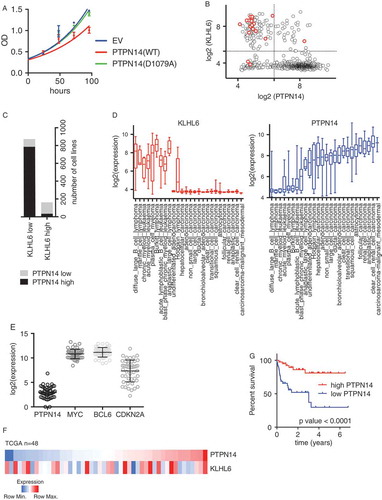
This is in accordance with the fact that a significant anti-correlation between PTPN14 and KLHL6 expression was observed in most cancer cell lines, as retrieved from public data available in the Cancer Cell Line Encyclopedia (CCLE) [Citation37](). Notably, majority of DLBCL cell lines expressing KLHL6 displayed low levels of PTPN14, suggesting that the two proteins might share the same downstream pathway. This correlation was particularly evident in lymphoid cancers ().
To confirm these observations in DLBCL patients, we analyzed the RNA-seq dataset from TCGA. The expression of PTPN14 is relatively low when compared to established oncogenes such as MYC and BCL6 as well as a tumor-suppressing gene CDKN2A (). As observed in DLBCL cells lines, the expression of KLHL6 and PTPN14 displayed an inverse correlation, although it did not reach a significant threshold due to the low number in the dataset ().
Finally, the low expression of PTPN14 correlated with significantly poorer survival rates in a cohort of DLBCL patients [Citation38] (), similar to the previous observations for KLHL6 [Citation7,Citation39,Citation40].
All together, our analysis revealed that PTPN14 is a down-regulated gene and a putative tumor suppressor with regard to DLBCL.
Discussion
Here, we show that tyrosine 691 in Roquin2 is phosphorylated in vivo. This event dissociates KLHL6 from Roquin2, thus inhibiting KLHL6-mediated ubiquitylation and degradation. Using proteomic to analyze Roquin2 complex, we identified the tyrosine phosphatase PTPN14. PTPN14 interacts with Roquin2 and dephosphorylates it at the tyrosine 691. Accordingly, PTPN14-mediated Roquin2 dephosphorylation impacts Roquin2 protein stability. A catalytic inactive PTPN14 mutant inhibits the degradation of Roquin2. Interestingly, our mapping revealed that the phosphatase domain of PTPN14 binds Roquin2 at the C-terminal. This complies with the fact that PTPN14 is specific to Roquin2. Indeed, Roquin1 and Roquin2 display high-sequence homology in the N-Terminal region but differ in the C-Terminus [Citation32], where the sequence similarity with Roquin1 is reduced.
It has been proposed that phosphorylation negatively impacts ligase-substrate pairing. For instance, phosphorylation of p85β has been proved to inhibit the interaction with its ligase FBXL2 [Citation41]. Furthermore, Aurora A is phosphorylated in serine residue within “A-box” that inhibits APC/C (the ubiquitin ligase anaphase-promoting complex)-dependent degradation [Citation42,Citation43].
PTPN14 has been regarded as a tumor suppressor in a variety of cancers as mutations and deletions are identified [Citation19–Citation21]. Consistent with this data, we found that PTPN14 negatively regulates the stability of Roquin2, a pro-proliferative gene in ABC-DLBCL cell lines [Citation7]. The role of protein tyrosine phosphatases in lymphoma has begun to be investigated. In Hodgkin lymphoma and primary mediastinal B cell lymphoma, PTPN1 frequently harbors loss of function mutations, suggesting a tumor suppressing role [Citation44]. Furthermore, inactivating mutations and epigenetic silencing of PTPN13 have been reported in non-Hodgkin’s and Hodgkin’s lymphoma [Citation45]. Accordingly, we have found that PTPN14 protein is expressed at very low levels in DLBCL cell lines, and the overexpression of PTPN14 decreases lymphoma cell proliferation. Since PTPN14 is deregulated in DLBCL patients, it is imperative to determine the molecular mechanism of PTPN14 down-regulation.
Finally, the finding that tyrosine 691 can be phosphorylated in cells suggests that a potential tyrosine kinases might be involved in this process. Recent study demonstrates that Roquin1 can specifically interact with the α1 subunit of AMPK kinase and negatively regulate AMPK kinase activity in follicular helper T cells [Citation46]. Furthermore, Roquin2 can interact and promote ubiquitylation and proteasomal degradation of ASK1 kinase upon the oxidative stress to inhibit reactive oxygen species (ROS)-induced cell death [Citation47]. Although, to date, there are no tyrosine kinases identified that associate with Roquin2 proteins, it is possible that PTM crosstalk between potential tyrosine kinase and PTPN14 phosphatase exists to fine-tune the regulation of Roquin2 stability, particularly in the context of BCR signaling. Finally, the Cyclin-depended kinase 1 (CDK1) has been recently reported as a KLHL6 substrate [Citation48]. Thus, it will be important to assess whether the mechanism of substrate regulation via phosphorylation is conserved amongst KLHL6 targets.
Methods
Cell lines, antibodies and reagents
HEK293T cells were cultured using Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS). OCI-LY10 cells were cultured using RPMI 1640 medium containing 10% FBS.
All antibodies were diluted with 1:1000 ratios unless otherwise specified. The following antibodies were used: anti-phospho tyrosine (Millipore Sigma, #05–321, 1:5000), anti-FLAG (Sigma, F7425, 1:3000), anti-Tubulin (sc-8035), anti-KLHL6 (ab182163), anti-CDK1 (sc-954), anti-CDK2 (sc-163), anti-PTPN14 (CST, #13,808), anti-V5 tag (CST, #13,202), anti-HA (Biolegend, #901,513), anti-Cullin1 (Invitrogen, #71–8700), anti-Roquin1/2 (MABF288). Anti-phospho Roquin2 Y691 antibody was generated with Yenzym Antibodies LLC using the peptide CPVPSGM-pY-APVYDSR. Anti-FLAG-M2 affinity gel (Sigma, A2220) and Strep-Tactin Superflow 50% suspension (Neuromics) were used for pull-down assays. 3X FLAG peptide (Sigma F4799) was used for FLAG-elution. Goat-F(ab’)2 anti-human IgM (SouthernBiotech, #2022–01; 10μg/ml final concentration) was used for BCR stimulation. The following drugs were used: Doxycycline hyclate (Sigma Aldrich; 1 μg/ml final concentration) and Puromycin (Sigma Aldrich; 0.5μg/ml-1μg/ml final concentration).
MTS assay was performed according to manufacture (Promega).
Plasmids and cloning
Dr. Carola Vinuesa kindly provided human Roquin1 and Roquin2 cDNA and we sub-cloned Roquins into different vectors: pcDNA3.1-FLAG, pcDNA3.1-FLAG-Streptavidin, PMSCV-FLAG-HA. Human KLHL6 cDNA was purchased from Dharmacon and we sub-cloned into pcDNA3.1-FLAG vector. The pcDNA3.1 V5-PTPN14 (#61,003) vector was purchased from Addgene and sub-cloned into pcDNA3.1-FLAG vector. Dr. Elizabeth White kindly provided all MSCV-N-term V5-tagged PTPN14 deletion mutants. cDNAs encoding HA-tagged KLHL6 was sub-cloned into MIGR1 retroviral vector. HA-tagged PTPN14 cDNA were sub-cloned into pTRIPZ vector. QuikChange Site-directed mutagenesis kit (Stratagene) was used to generate C-terminal deletion mutants and point mutants of Roquin2 and PTPN14. Standard PCR methods were used to generate PTPN14 PTP domain.
Transient transfections and generation of viruses for infection
HEK293T cells were transiently transfected using Polyethylenimine (PEI). For retrovirus and lenvirius production, GP-293 packaging cells (Clontech) or pCMV-DeltaR8.2 were used respectively. After 48 hours of transient transfection, the virus-containing medium was collected, filtered, and used for spin-infection at 1800 rpm for 30 minutes. Cells were incubated with virus supernatant plus 10 µg/ml of polybrene for six hours to overnight.
Immunoblotting and quantification of protein
Cell lysis was performed with RIPA-lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, 1% NP-40) supplemented with protease and phosphatase inhibitors and N-ethylmaleimide (Sigma Aldrich; 10µM). Protein concentrations were quantified using a Bio-Rad DC protein assay (Lowry assay) according to the manufacturer’s protocol. Equal amounts of protein (10-15μg) were loaded for SDS-PAGE (6% polyacrylamide gel) analyzes and confirmed by staining the membrane with Ponceau- S or immunoblotting with loading controls.
Immunoprecipitation
Cell lysis was performed with NP40 buffer (15 mM Tris pH 7.4, 1 mM EDTA, 150 mM NaCl, 1 mM MgCl2, 10% glycerol, 0.1% NP-40) containing protease inhibitors and DTT (1µM). After 5 min of lysis, the whole cell lysates were spun at 15,000 rpm for 5 min at 4 ºC. The supernatants were incubated with 15 μl of anti-FLAG resin or anti-streptavidin resin per each sample for 2 hrs at 4 ºC. After 4 washes with NP-40 lysis buffer, the immunoprecipitates were eluted with 2X Laemmli buffer (240 mM Tris pH 6.8, 8% SDS, 0.04% bromophenol blue, 5% β-mercaptoethanol, 40% glycerol) and boiled for electrophoresis.
In vitro phosphatase assay
FLAG-tagged phosphorylated-Roquin2, FLAG-tagged PTPN14 WT and FLAG-tagged PTPN14 PTP domain were immunoprecipitated by anti-FLAG-M2 affinity gel agarose beads from HEK293T cells. The immunocomplexes were washed 4 times with NP-40 lysis buffer, and twice in phosphatase assay buffer (50mM Tris-HCl PH 7.4, 150mM NaCl, 2µM DTT). The FLAG-tagged phosphorylated-Roquin2 immunocomplexes were incubated with beads alone, FLAG-tagged PTPN14 WT immunocomplexes, or FLAG-tagged PTPN14 PTP domain immunocomplexes in 40µl of phosphatase assay buffer plus 3µl of FLAG peptides (4mg/ml) for elution at 37ºC for 90 min in the thermo-mixer (1200 rpm). Immunoblot analysis was performed to quantitate phosphorylated Roquin2 at tyrosine 691.
Pervanadate treatment
Pervanadate mix (100mM sodium vandate, 100mM H2O2, ddH2O with 1:1:8 ratio) was made and incubated at room temperature for 5min. The cells were treated with pervanadate mix with a final concentration of 100 µM for 15min before cell collection.
Lamda(λ)-phosphatase treatment
After the fours washes of the immunoprecipitation protocol, the beads were washed two times with the Tris-NaCl buffer (25 mM Tris pH 7.4, 50mM NaCl) and incubated with the reaction mix (25 mM Tris pH 7.4, 50mM NaCl, 0.1 mM MnCl2, 1X NEB buffer PMP) with or without 1 µl of λ-phosphatase (New England Biolabs, P0753, 400 U) for 30 min at 30 ºC. The immunoprecipitates were eluted with 2X Laemmli buffer.
In vitro binding assay
In vitro-translated FLAG-tagged KLHL6 was incubated with indicated amounts of Roquin2 peptides (WT or phosphorylated form) in NP40-buffer (15 mM Tris pH 7.4, 1 mM EDTA, 150 mM NaCl, 1 mM MgCl2, 10% glycerol, 0.1% NP-40). Anti-streptavidin resin was added to the samples and incubated for 2hr with rotation at 4ºC. Samples were washed three times with the NP40-buffer, and protein complexes were eluted in Laemmli buffer.
Protein purification for mass spectrometry
HEK293T cells were transfected with FLAG-STREP-tagged Roquin1 or FLAG-STREP-tagged Roquin2 and lysed in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 0.5% NP40) with protease and phosphatase inhibitors. FLAG-STREP Roquin1 or Flag-STREP Roquin2 was immunoprecipitated with anti-FLAG agarose beads and washed five times with lysis buffer and proteins were eluted with FLAG peptides. 1% of protein eluates were separated by SDS-PAGE and stained by Silver Staining (Life Technology). The final eluates were precipitated with trichloroacetic acid (TCA) for mass spectrometry analysis.
MudPIT analysis
TCA-precipitated proteins were urea-denatured, reduced, alkylated and digested with endoproteinase Lys-C (Roche) and modified trypsin (Roche), as previously described [Citation49,Citation50]. Peptide mixtures were loaded onto 100-µm fused silica microcapillary columns packed with 5-μm C18 reverse phase (Aqua, Phenomenex), strong cation exchange particles (Luna, Phenomenex), and reverse phase [Citation51]. Loaded microcapillary columns were placed in-line with a Quaternary Agilent 1100 series HPLC pump and a LTQ linear ion trap mass spectrometer equipped with a nano-LC electrospray ionization source (Thermo Scientific). Fully automated 10-step MudPIT runs were performed on the electrosprayed peptides, as previously described [Citation49]. Tandem mass (MS/MS) spectra were analyzed using SEQUEST [Citation52] against a database of 61,318 sequences, consisting of 30,449 non-redundant human proteins (downloaded from NCBI on 2012–08-27, 160 usual contaminants (such as human keratins, IgGs and proteolytic enzymes), and, to estimate false discovery rates, 30,659 randomized amino-acid sequences derived from each non-redundant protein entry. Peptide/spectrum matches were sorted and selected using DTASelect with the following criteria set: spectra/peptide matches were only retained if they had a DeltCn of at least 0.08 and a minimum XCorr of 1.8 for singly-, 2.0 for doubly-, and 3.0 for triply-charged spectra. Additionally, peptides had to be fully tryptic and at least seven amino acids long. Combining all runs, proteins had to be detected by at least two such peptides, or one peptide with two independent spectra. Under these criteria the final FDRs at the protein and spectral levels were 2.1%± 0.3 and 0.94% ± 0.03, respectively. Peptide hits from multiple runs were compared via CONTRAST [Citation53]. Normalized Spectral Abundance Factors (NSAFs) were calculated for each detected protein, as previously described [Citation54–Citation56]. ftp://massive.ucsd.edu/MSV000082202.
Data availability
KLHL6 expression was calculated through re-analysis of microarray from KLHL6 and PTPN14 expression in cell line and patients were from CCLE and TCGA, respectively. Raw data for the proteomic analysis are available at http://www.stowers.org/research/publications/libpb-1289.
Author contributions
L.B. directed the project and oversaw the results. J.C. designed and performed all experiments. A.S., L.F. and M.P.W. performed the mass spectrometry analysis of the Roquin1/Roquin2 complex purified by J.C. L.B. and J.C. wrote the manuscript.
Supplemental Material
Download MS Excel (94.8 KB)Acknowledgments
The authors thank Dr. Carola Vinuesa for kindly providing Roquins cDNAs, and Dr. Elizabeth White for kindly providing MSCV-N-term V5-tagged PTPN14 deletion mutants. This work was supported in part by grant R00-CA166181-04, R01-CA207513-01 from the National Cancer Institute and Gilead Sciences Research Scholars Program in Hematology/Oncology to L.B.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary material data can be accessed here.
Additional information
Funding
References
- Nguyen, L.K, Muñoz-García J, Maccario H, et al. Switches, excitable responses and oscillations in the Ring1B/Bmi1 ubiquitination system. PLoS Comput Biol. 2011;7:e1002317.
- Moeller HB, Praetorius J, Rutzler MR, et al. Phosphorylation of aquaporin-2 regulates its endocytosis and protein-protein interactions. Proc Natl Acad Sci U S A. 2010;107:502–507.
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738.
- Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381.
- Bassermann F, Eichner R, Pagano M. The ubiquitin proteasome system - implications for cell cycle control and the targeted treatment of cancer. Biochim Biophys Acta. 2014;1843:446–453.
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479.
- Choi J, Lee K, Ingvarsdottir K, et al. Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2. Nat Cell Biol. 2018;20:586–596.
- Morin, R.D., Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298.
- Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884.
- Idoia GR, Kremer Hovinga JA, Terrell DR, et al. CREBBP loss cooperates with BCL2 over-expression to promote lymphoma in mice. Blood. 2016;128:2175–2178.
- Reddy, A., Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494. e415.
- Vinuesa CG, Cook MC, Angelucci C, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458.
- Leppek K, Schott J, Reitter S, et al. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153:869–881.
- Schlundt A, Heinz GA, Janowski R, et al. Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation. Nat Struct Mol Biol. 2014;21:671–678.
- Tan D, Zhou M, Kiledjian M, et al. The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nat Struct Mol Biol. 2014;21:679–685.
- Smith AL, Nukina N, Masuda N, et al. Pez: a novel human cDNA encoding protein tyrosine phosphatase- and ezrin-like domains. Biochem Biophys Res Commun. 1995;209:959–965.
- Wadham C, Gamble JR, Vadas MA, et al. The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates beta-catenin. Mol Biol Cell. 2003;14:2520–2529.
- Wadham C, Gamble JR, Vadas MA, et al. Translocation of protein tyrosine phosphatase Pez/PTPD2/PTP36 to the nucleus is associated with induction of cell proliferation. J Cell Sci. 2000;113(Pt 17):3117–3123.
- Zhang P, Giannini C, Sarkaria JN, et al. Identification and functional characterization of p130Cas as a substrate of protein tyrosine phosphatase nonreceptor 14. Oncogene. 2013;32:2087–2095.
- Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274.
- Wang, Z., Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166.
- Wang W, Gagliardini V, Raissig MT, et al. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 2012;26:1959–1971.
- Liu X, Giannini C, Sarkaria JN, et al. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene. 2013;32:1266–1273.
- Belle L, Ali N, Lonic A, et al. The tyrosine phosphatase PTPN14 (Pez) inhibits metastasis by altering protein trafficking. Sci Signal. 2015;8:ra18.
- Mello SS, Valente LJ, Raj N, et al. A p53 super-tumor suppressor reveals a tumor suppressive p53-Ptpn14-Yap axis in pancreatic cancer. Cancer Cell. 2017;32:460–473. e466.
- Heffetz D, Bushkin I, Dror R, et al. The insulinomimetic agents H2O2 and vanadate stimulate protein tyrosine phosphorylation in intact cells. J Biol Chem. 1990;265:2896–2902.
- Secrist JP, Burns LA, Karnitz L, et al. Stimulatory effects of the protein tyrosine phosphatase inhibitor, pervanadate, on T-cell activation events. J Biol Chem. 1993;268:5886–5893.
- Seet BT, Dikic I, Zhou -M-M, et al. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483.
- Sgromo A, Raisch T, Bawankar P, et al. A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin. Nat Commun. 2017;8:14307.
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440.
- Provost E, Weier CA, Leach SD. Multiple ribosomal proteins are expressed at high levels in developing zebrafish endoderm and are required for normal exocrine pancreas development. Zebrafish. 2013;10:161–169.
- Pratama A, Ramiscal R, Silva D, et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity. 2013;38:669–680.
- Ogata M, Takada T, Mori Y, et al. Effects of overexpression of PTP36, a putative protein tyrosine phosphatase, on cell adhesion, cell growth, and cytoskeletons in HeLa cells. J Biol Chem. 1999;274:12905–12909.
- White EA, Munger K, Howley PM. High-risk human papillomavirus E7 proteins target PTPN14 for degradation. MBio. 2016;7.
- Blanchetot C, Chagnon M, Dube N, et al. Substrate-trapping techniques in the identification of cellular PTP targets. Methods. 2005;35:44–53.
- Satpathy S, Wagner SA, Beli P, et al. Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol Syst Biol. 2015;11:810.
- Barretina J, Skaletsky H, Brown LG, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607.
- Lenz G, Link E, Parish S, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323.
- Kunder CA, Roncador G, Advani RH, et al. KLHL6 Is Preferentially Expressed in Germinal Center-Derived B-Cell Lymphomas. Am J Clin Pathol. 2017;148:465–476.
- Meriranta L, Kremer Hovinga JA, Terrell DR, et al. Low expression and somatic mutations of the KLHL6 gene predict poor survival in patients with activated b-cell like diffuse large B-cell lymphoma. Blood. 2016;128:2175–2178.
- Kuchay S, Duan S, Schenkein E, et al. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol. 2013;15:472–480.
- Littlepage LE, Ruderman JV, Kato K, et al. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 2002;16:2274–2285.
- Song L, Rape M, Vasta V, et al. Substrate-specific regulation of ubiquitination by the anaphase-promoting complex. Cell Cycle. 2011;10:52–56.
- Gunawardana J, Harris SR, Croucher NJ, et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat Genet. 2014;46:329–335.
- Ying J, Li H, Cui Y, et al. Epigenetic disruption of two proapoptotic genes MAPK10/JNK3 and PTPN13/FAP-1 in multiple lymphomas and carcinomas through hypermethylation of a common bidirectional promoter. Leukemia. 2006;20:1173–1175.
- Ramiscal RR, Bowman CA, Russell DA, et al. Attenuation of AMPK signaling by ROQUIN promotes T follicular helper cell formation. Elife. 2015;4:e08698.
- Maruyama T, Araki T, Kawarazaki Y, et al. Roquin-2 promotes ubiquitin-mediated degradation of ASK1 to regulate stress responses. Sci Signal. 2014;7:ra8.
- Ying M, Shao X, Jing H, et al. Ubiquitin-dependent degradation of CDK2 drives the therapeutic differentiation of AML by targeting PRDX2. Blood. 2018;131:2698–2711.
- Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–175.
- Washburn MP, Wolters D, Yates JR. 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247.
- McDonald WH, Ohi R, Miyamoto DT, et al. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom. 2002;219:245–251.
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989.
- Tabb DL, McDonald WH, Yates JR. 3rd DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26.
- Florens L, Carozza M, Swanson S, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311.
- Paoletti AC, Parmely TJ, Tomomori-Sato C, et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103:18928–18933.
- Zybailov B, Mosley AL, Sardiu ME, et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347.
