ABSTRACT
Background: Exosomes secreted by human mesenchymal stem cells (hMSCs) have been shown to promote cartilage regeneration. This study aimed to explore whether exosomal lncRNA-KLF3-AS1 derived from hMSCs can promote chondrocyte proliferation via miR-206/GIT1 axis in osteoarthritis (OA).
Methods: hMSCs and MSC-derived exosomes (MSC-exo) were prepared for morphological observation and identification by transmission electron microscopy (TEM) and flow cytometry. IL-1β-induced OA chondrocytes and collagenase-induced mouse OA model were established for the further experiments. Luciferase activity assay was performed to test whether miR-206 could bind to KLF3-AS1 or GIT1. Cell proliferation and apoptosis were evaluated by CCK-8 assay and flow cytometry, respectively.
Results: MSC-Exos increased chondrogenic genes Col2a1 (type II collagen alpha 1) and aggrecan, decreased hondrocyte hypertrophy markers MMP-13 (matrix metalloproteinase-13) and Runx2 (runt-related transcription factor 2) in chondrocytes isolated from OA model mice. Furthermore, MSC-Exos attenuated IL-1β-induced chondrocyte proliferation inhibition and apoptosis induction. Moreover, MSCKLF3-AS1-Exos (exosomes derived from KLF3-AS1-overexpressing-MSCs) ameliorated IL-1β-induced chondrocyte injury. Results also demonstrated that KLF3-AS1 acted as a competitive endogenous RNA (ceRNA) by sponging miR-206 to facilitate GIT1 expression. In addition, miR-206 overexpression and GIT1 knockdown reversed MSCKLF3-AS1-Exos-mediated attenuation of chondrocyte injury.
Conclusion: Exosomal KLF3-AS1 derived from MSCs involved in MSC-Exos-mediated chondrocyte proliferation induction and chondrocyte apoptosis inhibition via miR-206/GIT1 axis.
Abbreviation: G-protein-coupled receptor kinase interacting protein-1 (GIT1)
Introduction
Osteoarthritis (OA) is the most common form of joint disease, resulting in chronic pain, stiffness, and disability [Citation1]. It is characterized by progressive destruction of articular cartilage and cartilage loss caused by disequilibrium between anabolism and catabolism of chondrocytes and extracellular matrix [Citation2]. However, repair and regeneration of damaged articular cartilage is challenging due to the generally poor healing capacity of cartilage.
Mesenchymal stem cell (MSC) therapies have demonstrated efficacy in cartilage repair in both animal studies [Citation3] and human clinical trials [Citation4]. The efficacy of MSC-based therapies, which was previously predicated on the chondrogenic potential of MSC, has been increasingly attributed to the paracrine secretion, particularly exosomes. Exosomes, a subset of extracellular vesicles, are thought to function primarily as intercellular communication vehicles to transfer bioactive lipids, proteins, nucleic acids including mRNAs, microRNAs, and long noncoding RNAs (lncRNAs) between cells to elicit biological responses in recipient cells [Citation5,Citation6]. Recent evidence suggests that exosomes secreted by human mesenchymal stem cells (MSC-Exos) have the capacity of promoting cartilage regeneration [Citation7,Citation8], however, the underlying mechanisms are not fully understood.
MicroRNAs (miRNAs) are evolutionarily conserved, endogenous non-coding RNAs of about 22 nucleotides that post-transcriptionally regulate gene expression through binding to the 3ʹ-untranslated region (3ʹ-UTR) of target mRNAs, leading to translational repression or target degradation [Citation9]. miRNAs play crucial roles in all cellular processes and also important in the process of chondrogenesis and cartilage remodeling [Citation10]. A recent study [Citation11] demonstrated that miR-206, which was significantly increased in human OA tissues and chondrocytes, significantly inhibited the proliferation and promoted apoptosis of chondrocytes. These data indicated that miR-206 may involve in cartilage degradation in OA and manipulation of miR-206 expression in human chondrocytes may be a novel therapeutic strategy for OA.
Extensive research has shown that G-protein-coupled receptor kinase interacting protein-1 (GIT1) promotes chondrocytes proliferation and inhibits chondrocytes apoptosis [Citation12]. Zhao et al. [Citation13] stated that platelet-derived growth factor (PDGF) promotes chondrocyte proliferation but suppresses chondrocyte apoptosis via upregulating GIT1 expression. A recent study also showed that [Citation14] miR-195 inhibits the proliferation and migration of chondrocytes by targeting GIT1.
A novel lncRNA KLF3-AS1 (KLF3 Antisense RNA 1; Ensembl: ENST00000440181) from MSC-Exos has been previously reported in ExoCarta, a manually web-based database (http://www.exocarta.org) of the exosomal molecular components. Nevertheless, the potential role of exosomal KLF3-AS1 in OA has not yet been elucidated. KLF3-AS1 harbors putative binding sites of miR-206 using DIANA TOOLS. Furthermore, GIT1 was identified as a putative target of miR-206 by harboring a miR-206 binding sequence in the 3ʹ-UTR of its mRNA (Targetscan). LncRNAs have been well known to exert roles by acting as a competitive endogenous RNA (ceRNA) to segregate microRNAs (miRNAs) away from target mRNAs [Citation15–Citation17]. Therefore, we speculated that, exosomal KLF3-AS1 derived from MSCs might act as a ceRNA by sponging miR-206 to facilitate GIT1 expression, and thereby promote chondrocyte proliferation and inhibit chondrocyte apoptosis. To address this, we detected MSC-derived exosomal KLF3-AS1 expression. Furthermore, the interaction among KLF3-AS1, miR-206, and GIT1 was investigated. Moreover, the role of exosomal KLF3-AS1/miR-206/GIT1 axis in regulating the function of chondrocytes in OA was also explored.
Results
Morphological observation and identification of MSCs and MSC-Exos
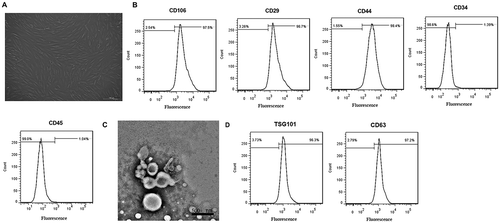
After 72 h of culture, human MSCs (hMSCs) showed uniform morphology and spindle-shaped appearance and were arranged in whorls (()). Furthermore, flow cytometry displayed that almost all of the cells were positive for classical MSC surface markers CD106, CD29 and CD44, and negative for hematopoietic lineage and endothelial markers CD34 and CD45 (()), indicating these cells belonged to high-purity MSCs. MSC-Exos identification was also performed by TEM as well as flow cytometry. As shown in (), the yielded vesicles were consistent with exosomes in size and morphology (round-shaped). Moreover, flow cytometry analysis showed that MSC-Exos were positive for the exosomal surface markers TSG101 and CD63 (()). These results indicated that the obtained MSC-Exos confirmed to the well-established criteria.
Figure 1. Morphological observation and identification of MSCs and MSC-Exo. (a) Phase-contrast microscopy of human bone marrow-derived MSCs in the third passage (scale bar: 100 μm). (b) Analysis of the immunophenotype of mouse bone marrow-derived MSCs by flow cytometry. MSCs were positive for CD29, CD106, and CD44, while negative for CD34, CD45. (c) Morphology of MSC-derived exosomes (MSC-Exos) under an Transmission electron microscope. Scale bar, 500 nm. (d) MSC-Exos were positive for TSG101 and CD63 as indicated by flow cytometry analysis.
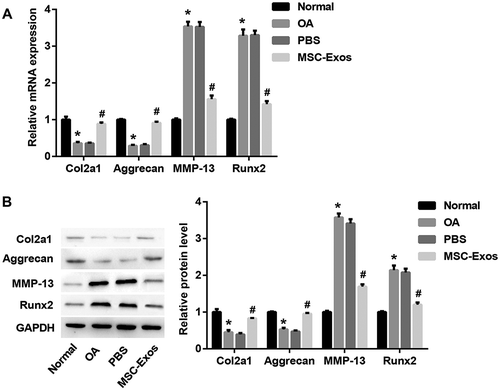
MSC-Exos increase Col2a1 and aggrecan, decrease MMP-13 and Runx2
Col2a1 (type II collagen alpha 1) and aggrecan are major cartilage extracellular matrix proteins that are essential for normal cartilage function. Runx2 (runt-related transcription factor 2) is a transcription factor involved in chondrocyte differentiation and hypertrophy, and type II collagen-degrading MMP-13 (matrix metalloproteinase-13) contributes to cartilage degradation [Citation18]. To determine the role of MSC-Exos in chondrocytes, we detected the levels of these key cartilage genes with MSC-Exos treatment in OA mice-derived chondrocytes. Data indicated that chondrogenic genes Col2A1 and aggrecan were notably downregulated, whereas chondrocyte hypertrophy markers MMP-13 and Runx2 were markedly upregulated, at both mRNA (()) and protein (()) levels, in chondrocytes isolated from OA model mice as compared with the chondrocytes isolated from normal mice. Interestingly, MSC-Exos treatment led to a significant increase in Col2A1 and aggrecan, and a notable decrease in MMP-13 and Runx2 ((,)), indicating the potential role of MSC-Exos in inhibiting chondrocytes degradation.
Figure 2. MSC-Exos increased Col2a1 and aggrecan, decreased MMP-13 and Runx2. The chondrocytes were divided into four groups: normal group (chondrocytes isolated from normal mice), OA group (chondrocytes isolated from OA model mice), MSC-Exos group (10 μg/ml MSC-Exos were added into OA mice-derived chondrocytes) and PBS group (as control of MSC-Exos). The relative mRNA (a) and protein (b) levels of Col2a1, aggrecan, MMP-13, and Runx2 were examined by qRT-PCR and Western blot, respectively. GAPDH served as the loading control. *p < 0.05 vs. the normal group; #p < 0.05 vs. the PBS group.
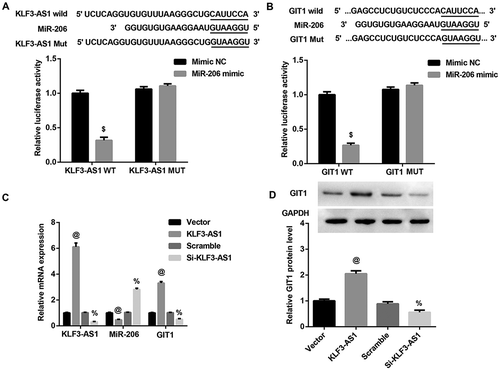
KLF3-AS1 acts as a ceRNA by sponging miR-206 to facilitate GIT1
We next investigated the interaction among KLF3-AS1, miR-206, and GIT1. Luciferase reporter assay showed that miR-206 mimic led to a marked decrease in luciferase activity in KLF3-AS1-WT reporter compared with the mimic NC group, whereas had no obvious effect on the luciferase activity in KLF3-AS1-MUT reporter, verifying the direct binding between KLF3-AS1 and miR-206 (()). Moreover, we also observed decreased luciferase activity in HEK293T cells co-transfected with WT GIT1 3ʹUTR luciferase reporter plasmids and miR-206 mimics (()), suggesting that 3ʹUTR of GIT1 is directly targeted by miR-206. In addition, KLF3-AS1 overexpression significantly inhibited miR-206 expression, whereas induced GIT1 expression at both mRNA (()) and protein (()) levels. In contrast, KLF3-AS1 knockdown exerted the opposite effects. Collectively, these results indiated that KLF3-AS1 acted as a sponge for miR-206 to facilitate GIT1 expression.
Figure 3. KLF3-AS1 acts as a ceRNA by sponging miR-206 to facilitate GIT1. (a) Results of luciferase activity assay verified the direct binding between KLF3-AS1 and miR-206. $p < 0.05 vs. the mimic NC+ KLF3-AS1 WT group. (b) Results of luciferase activity assay showed that 3ʹUTR of GIT1 is directly targeted by miR-206. $p < 0.05 vs. the mimic NC+ GIT1 WT group. (c-d) KLF3-AS1 overexpression significantly inhibited miR-206 expression, whereas induced GIT1 expression at both mRNA (c) and protein (d) levels. In contrast, KLF3-AS1 knockdown exerted the opposite effects. @p < 0.05 vs. the vector group; %p < 0.05 vs. the scramble group.
Exosomal KLF3-AS1 derived from MSCs attenuates IL-1β-induced chondrocyte injury
We next explored whether MSC-Exos exerted therapeutical effects on interleukin-1β (IL-1β)-induced chondrocyte injury via exosomal KLF3-AS1 derived from MSCs. To address it, chondrocytes from normal C57BL/6 mice were treated with either normal medium, IL-1β, PBS, MSC-Exos, or MSCKLF3-AS1-Exos (exosomes derived from KLF3-AS1 overexpressing-MSCs). PCR revealed that KLF3-AS1 expression was detected in both MSCs and MSC-Exos (()). qRT-PCR results confirmed the efficiency of KLF3-AS1 overexpression in MSCs (()). Furthermore, consistent with results in OA mice-derived chondrocytes, increased miR-206 expression (()), decreased mRNA and protein expression of GIT1 ((,)), decreased Col2A1 and aggrecan, as well as increased MMP-13 and Runx2 ((,)) were also detected in IL-1β-induced chondrocytes. In addition, IL-1β treatment significantly inhibited cell proliferation (()) and induced cell apoptosis (()).
Figure 4. Exosomal KLF3-AS1 derived from MSCs attenuated IL-1β-induced chondrocyte injury. (a) PCR revealed that KLF3-AS1 expression can be detected in both MSCs and MSC-Exos. (b) qRT-PCR results confirmed the overexpression efficiency of KLF3-AS1 expression in MSCs. $p < 0.05 vs. the vector group. (c-d) Normal mouse chondrocytes were prestimulated by IL-1β (10 mg/ml, 24 h, normal medium as control) before being co-cultured with MSCvector-Exos, MSCKLF3-AS1-Exos, or equal volume of PBS as control for 24 h. Results of qRT-PCR and Western blot revealed that MSCvector-Exos treatment significantly reversed IL-1β-mediated effects on expression of miR-206, GIT1 (c-d), Col2A1, aggrecan, MMP-13, Runx2 (e-f). (G-H) Results of CCK-8 assay and flow cytometry showed that MSCvector-Exos treatment significantly reversed the IL-1β-mediated chondrocyte proliferation inhibition and apoptosis induction. MSCKLF3-AS1-Exos treatment further enhanced the MSCvector-Exos-mediated effects. *p < 0.05 vs. the control group; #p < 0.05 vs. the PBS group; $p < 0.05 vs. the MSCvector-Exos group.
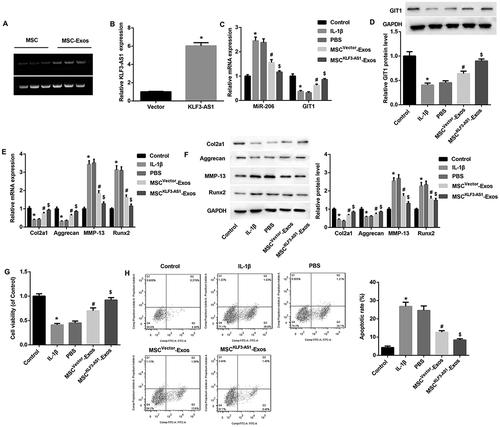
Importantly, MSCvector-Exos treatment significantly reversed the IL-1β-mediated unfavorable effects on chondrocytes in vitro, indicating that MSC-Exos alleviated IL-1β-induced OA chondrocyte injury by promoting chondrocyte proliferation and inhibiting apoptosis. Interestingly, MSCKLF3-AS1-Exos treatment further enhanced the MSCvector-Exos-mediated alleviation on chondrocyte injury ((-)). These data highlighted the importance of exosomal KLF3-AS1 derived from MSCs in the attenuation of IL-1β-induced chondrocyte injury.
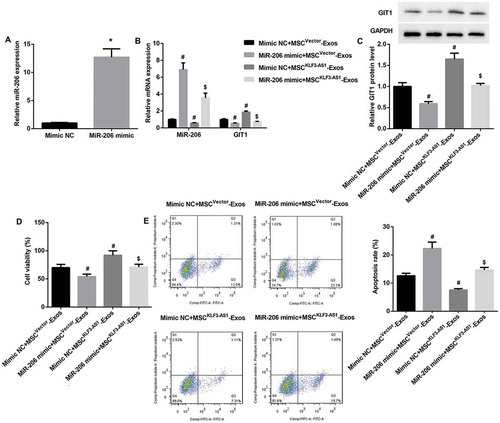
miR-206 overexpression reverses MSCKLF3-AS1-Exos-mediated attenuation of chondrocyte injury
To further investigate the role of KLF3-AS1/miR-206/GIT1 axis in MSC-Exos-mediated attenuation of IL-1β-induced chondrocyte injury, mouse chondrocytes were transfected with miR-206 mimic, after which cells were pre-stimulated by IL-1β before being co-cultured with MSCvector-Exos or MSCKLF3-AS1-Exos. qRT-PCR results confirmed that miR-206 expression was significantly upregulated in miR-206 mimic-transfected chondrocytes (()). Furthermore, miR-206 mimic significantly reversed MSCKLF3-AS1-Exos-mediated decrease in miR-206 expression and increase in mRNA and protein expression of GIT1 ((,)). Moreover, miR-206 mimic significantly reversed MSCKLF3-AS1-Exos-mediated chondrocyte proliferation induction (()) and chondrocyte apoptosis inhibition (()). Collectively, these results indicated that miR-206 overexpression in chondrocytes reversed MSCKLF3-AS1-Exos-mediated GIT1 upregulation, chondrocyte proliferation induction and apoptosis inhibition.
Figure 5. miR-206 overexpression reversed MSCKLF3-AS1-Exos-mediated attenuation of chondrocyte injury. (a) qRT-PCR results confirmed that miR-206 expression was significantly upregulated in miR-206 mimic-transfected chondrocytes. *p < 0.05 vs. the mimic NC group. The miR-206 mimic-transfected normal mouse chondrocytes were prestimulated by IL-1β (10 mg/ml, 24 h) before being co-cultured with MSCvector-Exos or MSCKLF3-AS1-Exos. (b-c) Results of qRT-PCR and Western blot revealed that miR-206 mimic significantly reversed MSCKLF3-AS1-Exos-mediated decrease in miR-206 expression and increase in mRNA and protein expression of GIT1. (d-e) Results of CCK-8 assay and flow cytometry showed that miR-206 mimic significantly reversed MSCKLF3-AS1-Exos-mediated cell proliferation induction and cell apoptosis inhibition. #p < 0.05 vs. the mimic NC + MSCvector-Exos group; $p < 0.05 vs. the mimic NC + MSCKLF3-AS1-Exos group.
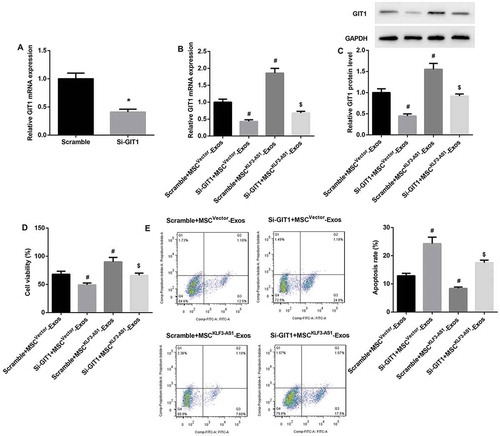
GIT1 knockdown reverses MSCKLF3-AS1-Exos-mediated attenuation of chondrocyte injury
Next, mouse chondrocytes were transfected with si-GIT1, after which cells were pre-stimulated by IL-1β before being co-cultured with MSCvector-Exos or MSCKLF3-AS1-Exos. qRT-PCR results confirmed that GIT1 mRNA expression was significantly downregulated in si-GIT1-transfected chondrocytes (()). Furthermore, GIT1 knockdown significantly reversed MSCKLF3-AS1-Exos-mediated increase in mRNA and protein expression of GIT1 ((,)). We also found that GIT1 knockdown significantly reversed MSCKLF3-AS1-Exos-mediated cell proliferation induction (()) and cell apoptosis inhibition (()). Together, these aforementioned results indicated that exosomal KLF3-AS1 derived from MSCs involved in MSC-Exos-mediated chondrocyte proliferation induction and chondrocyte apoptosis inhibition via miR-206/GIT1 axis.
Figure 6. GIT1 knockdown reversed MSCKLF3-AS1-Exos-mediated attenuation of chondrocyte injury. (a) qRT-PCR results confirmed that GIT1 mRNA expression was significantly downregulated in si-GIT1-transfected chondrocytes. *p < 0.05 vs. the scramble group. The si-GIT1-transfected normal mouse chondrocytes were prestimulated by IL-1β (10 mg/ml, 24 h) before being co-cultured with MSCvector-Exos or MSCKLF3-AS1-Exos. (b-c) Results of qRT-PCR and Western blot revealed that GIT1 knockdown significantly reversed MSCKLF3-AS1-Exos-mediated increase in mRNA and protein expression of GIT1. (d-e) Results of CCK-8 assay and flow cytometry showed that GIT1 knockdown significantly reversed MSCKLF3-AS1-Exos-mediated cell proliferation induction and cell apoptosis inhibition. #p < 0.05 vs. the scramble + MSCvector-Exos group; $p < 0.05 vs. the scramble + MSCKLF3-AS1-Exos group.
Discussion
Chondrocyte proliferation promotion and apoptosis inhibition have been the key strategies to prevent and control OA [Citation19,Citation20]. The present study demonstrated that MSC-Exos promoted proliferation but inhibited apoptosis of chondrocytes. Furthermore, the therapeutical effect of MSC-Exos on OA was related to newly discovered exosomal lncRNA KLF3-AS1, which acted as ceRNA by sponging miR-206 to facilitate GIT1 expression in chondrocytes.
To date, MSC-Exos have been shown to protect against myocardial ischemia/reperfusion injury [Citation21], ameliorate graft-versus-host disease [Citation22], enhance would healing [Citation23], and more recently improve cartilage and bone regeneration [Citation7,Citation8]. In this study, we observed that exosomes derived from hMSCs overturned downregulation of chondrogenic genes Col2A1 and aggrecan, upregulation of hypertrophy markers MMP-13 and Runx2 in OA chondrocytes and IL-1β-induced chondrocytes. Furthermore, MSC-Exos reversed IL-1β-induced inhibition of chondrocyte proliferation and promotion of chondrocyte apoptosis. These findings confirmed our speculation that MSC-Exos exerted therapeutical effects on OA.
In recent years, increasing studies provided strong evidence that MSC-Exos involve in damage repair process by delivering functional proteins or RNAs. For example, Wang et al. [Citation24] showed that exosomes derived from human umbilical cord MSCs improved myocardial repair via Smad7 upregulation. Tao et al. [Citation25] observed that exosomes derived from miR-140-5p-overexpressing synovial MSCs promoted the proliferation and migration of rat chondrocytes, which in turn ameliorated OA. In this study, by searching the differently expressed RNAs in MSC-Exos, we noted a novel lncRNA KLF3-AS1, which localizes at chromosome 4p14 and is enriched in MSC-Exos according to the exocarta database. The further PCR analysis confirmed the existence of lncRNA KLF3-AS1 in hMSCs and MSC-Exos. Importantly, exosomes derived from KLF3-AS1-overexpressing-MSCs further enhanced MSCvector-Exos-mediated amelioration of IL-1β-induced chondrocyte injury, suggesting the involvement of exosomal KLF3-AS1 in the therapeutical effect of MSC-Exos on OA.
LncRNAs have been well known to exert roles by acting as a competitive endogenous RNA (ceRNA) to segregate microRNAs (miRNAs) away from target mRNAs [Citation15–Citation17]. Using bioformatics analysis, we noted that KLF3-AS1 harbors putative binding sites of miR-206. Furthermore, GIT1 was identified as a putative target of miR-206 by harboring a miR-206 binding sequence in the 3ʹ-UTR of its mRNA. A recent study [Citation11] demonstrated that miR-206 overexpression significantly inhibited the proliferation, promoted chondrocyte apoptosis, dramatically decreased Col2a1 and aggrecan, and increased Runx2 and MMP-13 [Citation11], indicating the involvement of miR-206 in cartilage degradation in OA. Furthermore, GIT1 has been shown to promote chondrocytes proliferation and inhibit chondrocytes apoptosis [Citation12–Citation14]. In the current study, we verified not only the direct binding between KLF3-AS1 and miR-206, but also 3ʹUTR of GIT1 and miR-206. Furthermore, we observed that KLF3-AS1 acted as a ceRNA by sponging miR-206 to facilitate GIT1 expression. In addition, miR-206 overexpression and GIT1 knockdown reversed MSCKLF3-AS1-Exos-mediated attenuation of IL-1β-induced chondrocyte injury. These data indicated that the potential therapeutic effects of exosomal KLF3-AS1 derived from MSCs in OA is related to its segregation of miR-206 away from target GIT1, which in turn promotes chondrocytes proliferation and inhibits chondrocytes apoptosis.
In conclusion, MSCs derived exosomal lncRNA KLF3-AS1 transplantation could facilitate cartilage repair by promoting chondrocyte proliferation and inhibiting apoptosis via miR-206/GIT1 axis. Therefore, our findings highlighted the possible mechanism for OA therapy by cellular delivery of exosomal lncRNA KLF3-AS1.
Materials and methods
MSCs culture and identification
hMSCs were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco’s modified Eagles medium (DMEM)/F12 (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) in a humidified atmosphere containing 95% air and 5% CO2 at 37°C. The third passage hMSCs was used in subsequent experiments. The morphological characteristics of hMSCs were examined using an inverted microscope. Analysis of the immunophenotype (CD29, CD106, CD44, CD34, CD45, 1:100; BD Biosciences, San Jose, CA, USA) of hMSCs was analyzed on a FACScan flow cytometer (BD Biosciences).
Extraction and identification of MSC-derived exosomes
MSCs-conditioned medium was collected after 48 h, followed by centrifugation at 3,000g for 15 min. The media was incubated with 0.5 volume of total exosome isolation reagent (Thermo Scientific, Waltham, MA, USA) overnight at 4°C. The mixture was centrifuged at 12,000 g for 1 h and the supernatant was discarded. Exosome pellets were resuspended in PBS and prepared for identification.
The protein concentration of the MSC-Exos preparations was quantified using the Bradford method (Bio-Rad, Hercules, USA). The morphologic characteristics of MSC-Exos were observed by transmission electron microscopy (TEM). The phenotypic profile of MSC-Exos was determined by flow cytometry. Briefly, after washed with 0.1% BSA for three times, exosomes-coated beads were incubated with anti-TSG101 (1:500, Abcam, Cambridge, UK) and anti-CD63 (1:200, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) for 30 min, followed by incubation with anti-IgG secondary antibody (1:1000; Cell Signaling Technology, Boston, MA, USA) and analyzed on a FACSCalibur flow cytometer (BD Biosciences).
Establishment of OA model
Adult pathogen-free C57BL/6 mice (18–24 g; 9 weeks old) were obtained from Charles River Laboratories (Beijing, China). The experimental procedures were approved by the Animal Care and Use Committee of Luhe People’s Hospital of Nanjing. All mice were housed in a well-ventilated holding room with free access to food and water, and a 12/12 h light/dark cycle, with an ambient temperature of 20–25°C.
For the establishment of experimental OA mice model, type II collagenase solution (Worthington Biochemical, Lakewood, NJ, USA) was dissolved in sterile PBS (pH 7.4), filtered and then injected into the knee joint cavity of C57BL/6 mice under anesthesia.
Isolation and culture of chondrocytes
Chondrocytes were isolated from the knee joint of C57BL/6 mice. Briefly, the articular cartilage tissue from the right knee joint of the hindlegs of OA mice was cut into small pieces (< 1 mm [Citation3]) and incubated with 0.2% trypsin at 37°C for 30 min. Following removal of the trypsin solution, the tissue was treated with 2 ml 0.2% Type II collagenase at 37°C for 2 h. Released cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin (100 g/ml). The medium was replaced every 2 days. Following the growth of cells to 80–90% confluence, the third passage chondrocytes were used for all subsequent experiments.
Cell transduction and transfection
Lentiviral vectors that stably overexpressing KLF3-AS1, lentiviral vectors that stably silencing KLF3-AS1, siRNA targeting GIT1 (si-GIT1), miR-206 mimic, and their corresponding controls were purchased from GenePharma (Shanghai, China), and transfected/transduced into MSCs using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the overexpression or knockdown efficiency was examined by qRT-PCR.
Luciferase activity assay
The fragments of KLF3-AS1 and 3ʹ-untranslated region (UTR) of GIT1 containing the predicted wild-type (WT) binding sites of miR-206 or mutated miR-206 binding sites (MUT) were amplified by PCR and inserted into a pMIR-REPORT luciferase reporter vector (Ambion, Austin, TX, USA), named as KLF3-AS1-WT, KLF3-AS1-MUT, GIT1-WT, and GIT1-MUT. When the luciferase reporter assay is performed, cells were co-transfected with 200 ng constructed luciferase reporter vectors, 25 ng pRL-TK (expressing renilla luciferase as the internal control) and 20 μM miR-206 mimic or miR-negative control using Lipofectamine 2000. A luciferase reporter assay system (Promega Corporation, Fitchburg, WI, USA) was applied to analyze the luciferase activities at 24 h post transfection.
Cell proliferation assay
Chondrocyte growth was measured using the cell counting kit-8 (CCK-8, Sigma, USA) according to the manufacturer’s protocol. Briefly, when the confluence reached 70%-80%, 10 µl CCK-8 solution was added into each well and incubated at 37°C for 3 h. The optical density (OD) at 450 nm was measured with averages from six replicates using a microplate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Cell apoptosis assay
An annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) cell apoptosis kit was used according to the instructions (Invitrogen, Thermo Fisher Scientific, Inc.) to quantify chondrocyte apoptosis. Briefly, chondrocytes (4 × 105) were washed twice with PBS buffer (pH 7.4), and re-suspended in the binding buffer provided in the kit. After this, 5 µl Annexin V-FITC and 5 µl PI were mixed with the cells. Following 15 min of incubation at room temperature in the dark, mixtures were analyzed using the FACScan flow cytometry (BD Biosciences).
Real time quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from chondrocytes or mouse knee cartilage using TRIzol kit (Invitrogen). RNA was then reverse transcribed into cDNAs using the iScript cDNA synthesis kit (Bio-rad, Germany) according to the manufacturer’s instructions. RT-qPCR was carried out using the SYBR® Premix Ex TaqTM kit (Takara Bio, Inc., Otsu, Japan), according to the manufacturer’s instructions. The primers used were as follows: KLF3-AS1: F: 5ʹ- AATGAGTGCGTGGAGGAAAT −3ʹ, R; 5ʹ- CCTGGGCAACAGAGTGAGAC-3ʹ; miR-206: F: 5ʹ- GGAATGTAAGGAAGTGTGTG-3ʹ, R: 5ʹ- GAACATGTCTGCGTATCTC-3ʹ; GIT1: F: 5ʹ- GGTCACCGCCAAAGACCTC-3ʹ, R: 5ʹ- CAGCCCCATACACTACCAGC-3ʹ; Col2a1: F: 5ʹ- GGGAATGTCCTCTGCGATGAC-3ʹ, R: 5ʹ- GAAGGGGATCTCGGGGTTG −3ʹ; aggrecan: F: 5ʹ- CCTGCTACTTCATCGACCCC −3ʹ, R: 5ʹ- AGATGCTGTTGACTCGAACCT −3ʹ; MMP-13: F: 5ʹ- GATGACCTGTCTGAGGAAGACC-3ʹ, R: 5ʹ- GCATTTCTCGGAGCCTGTCAAC-3ʹ; Runx2: F: 5ʹ- AACGATCTGAGATTTGTGGGC-3ʹ, R: 5ʹ- CCTGCGTGGGATTTCTTGGTT-3ʹ. The thermocycling conditions used were as follows: 95°C for 3 min; followed by 40 cycles of 95°C for 5 sec, 60°C for 30 sec and 72°C for 30 sec, followed by a final extension of 72°C for 5 min. Relative expression of these genes were calculated by the 2−ΔΔCt method. Relative KLF3-AS1 and miR-206 expression were normalized to 18S, and the mRNA levels of other genes were normalized to GAPDH.
Western blot
The proteins isolated from chondrocytes or mouse knee cartilage were lysed in RIPA buffer (Thermo Scientific, USA) containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% SDS, 1% sodium deoxycholate, 1 mM EDTA, 1% Triton X-100 and protease inhibitors, according to the manufacturer’s protocol. The protein extraction was performed at 4°C for 30 min. The protein concentration in samples was measured using the Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). Proteins (20 μg/lane) were separated by 10% SDS-PAGE gels. Following electrophoresis, proteins were transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After being blocked with 5% fat-free milk, primary antibodies against Col2a1 (1:500, Santa Cruz Biotechnology Inc.), aggrecan (1:100, Santa Cruz Biotechnology Inc.), MMP13 (1:1000, Santa Cruz Biotechnology Inc.), Runx2 (1:200, Santa Cruz Biotechnology Inc.), GAPDH (1:1000; Santa Cruz Biotechnology Inc.) were added for incubation overnight at 4°C, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2000; Santa Cruz Biotechnology Inc.). GAPDH was used as the loading control. The protein was detected with an enhanced chemiluminescence kit (Applygen Technologies, China) and the band intensity was quantified with Image-Pro Plus 6.0 software.
Statistical analysis
All statistical analyses were performed using SPSS statistical software package standard version 16.0 (SPSS, Inc., Chicago, IL, USA). The data are presented as the mean ± standard deviation (SD) from three independent experiments. The unpaired Student’s t-test was used to analyze differences between two groups. One-way analysis of variance (ANOVA) was used to analyze differences among multiple groups. p < 0.05 was considered to indicate a statistically significant difference.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cross M, Smith E, Hoy D, et al. The global burden of hip and knee osteoarthritis: estimates from the global burden of disease 2010 study. Ann Rheum Dis. 2014;73:1323–1330.
- Fellows CR, Matta C, Mobasheri A. Applying proteomics to study crosstalk at the cartilage-subchondral bone interface in osteoarthritis: current status and future directions. Ebiomedicine. 2016;11:2–4.
- Guo W, Zheng X, Zhang W, et al. Mesenchymal stem cells in oriented PLGA/ACECM composite scaffolds enhance structure-specific regeneration of hyaline cartilage in a rabbit model. Stem Cells Int. 2018;2018:6542198.
- Kamei N, Ochi M, Adachi N, et al. The safety and efficacy of magnetic targeting using autologous mesenchymal stem cells for cartilage repair. Knee Surg Sports Traumatol Arthrosc. 2018.
- Toh WS, Lai RC, Po Hui JH, et al. MSC exosome as a cell-free MSC therapy for cartilage regeneration: implications for osteoarthritis treatment. Semin Cell Dev Biol. 2017;67:56.
- Li B, Xu H, Han H, et al. Exosome-mediated transfer of lncRUNX2-AS1 from multiple myeloma cells to MSCs contributes to osteogenesis. Oncogene. 2018.
- Qi X, Zhang J, Yuan H, et al. Exosomes secreted by human-induced pluripotent stem cell-derived mesenchymal stem cells repair critical-sized bone defects through enhanced angiogenesis and osteogenesis in osteoporotic rats. Int J Biol Sci. 2016;12:836.
- Zhang S, Chu WC, Lai RC, et al. Exosomes derived from human embryonic mesenchymal stem cells promote osteochondral regeneration. Osteoarthritis Cartilage. 2016;24:2135–2140.
- Xu W, Hang M, Yuan C-Y, et al. MicroRNA-139-5p inhibits cell proliferation and invasion by targeting insulin-like growth factor 1 receptor in human non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:3864–3870.
- Zhang Y, Jia J, Yang S, et al. MicroRNA-21 controls the development of osteoarthritis by targeting GDF-5 in chondrocytes. Exp Mol Med. 2014;46:e79.
- Ni Z, Shang X, Tang G, et al. Expression of miR-206 in human knee articular chondrocytes and effects of miR-206 on proliferation and apoptosis of articular chondrocytes. Am J Med Sci. 2018;355:240–246.
- Zhang LQ, Zhao GZ, Xu XY, et al. Integrin-β1 regulates chondrocyte proliferation and apoptosis through the upregulation of GIT1 expression. Int J Mol Med. 2015;35:1074–1080.
- Zhao G, Zhang L, Liu Y, et al. Effects of platelet-derived growth factor on chondrocyte proliferation, migration and apoptosis via regulation of GIT1 expression. Mol Med Rep. 2016;14:897–903.
- Gu Y, Rong X, Wen L, et al. miR-195 inhibits the proliferation and migration of chondrocytes by targeting GIT1. Mol Med Rep. 2017;15:194–200.
- Wang P, Chen D, Ma H, et al. LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis. Oncotargets Ther. 2017;10:5137.
- Xiong D, Li Z, Liang L, et al. The LncRNA NEAT1 accelerates lung adenocarcinoma deterioration and binds to Mir-193a-3p as a competitive endogenous RNA. Cell Physiol Biochem. 2018;48:905–918.
- Gu J, Wang Y, Wang X, et al. Effect of the LncRNA GAS5-MiR-23a-ATG3 axis in regulating autophagy in patients with breast cancer. Cell Physiol Biochem. 2018;48:194–207.
- Xu R, Li J, Wei B, et al. MicroRNA-483-5p modulates the expression of cartilage-related genes in human chondrocytes through down-regulating TGF-beta1 expression. Tohoku J Exp Med. 2017;243:41–48.
- Jin L, Zhao J, Jing W, et al. Role of miR-146a in human chondrocyte apoptosis in response to mechanical pressure injury in vitro. Int J Mol Med. 2014;34:451.
- Yan S, Wang M, Zhao J, et al. MicroRNA-34a affects chondrocyte apoptosis and proliferation by targeting the SIRT1/p53 signaling pathway during the pathogenesis of osteoarthritis. Int J Mol Med. 2016;38:201–209.
- Lai RC, Arslan F, Lee MM, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–222.
- Kordelas L, Rebmann V, Ludwig AK, et al. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia. 2014;28:970–973.
- Zhang J, Guan J, Niu X, et al. Exosomes released from human induced pluripotent stem cells-derived MSCs facilitate cutaneous wound healing by promoting collagen synthesis and angiogenesis. J Transl Med. 2015;13:49.
- Wang XL, Zhao YY, Sun L, et al. Exosomes derived from human umbilical cord mesenchymal stem cells improve myocardial repair via upregulation of Smad7. Int J Mol Med. 2018;41:3063–3072.
- Tao SC, Yuan T, Zhang YL, et al. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics. 2017;7:180.
