ABSTRACT
Increasing evidence showed that circular RNAs (circRNAs) play critical roles in tumorigenesis. However, the roles and underlying mechanisms of circRNAs in clear cell renal cell carcinoma (ccRCC) remain unclear. In the present study, we identified a novel circRNA circPCNXL2, which was significantly upregulated in ccRCC by circular RNA microarray. Further analysis revealed that circPCNXL2 was significantly increased and correlated with poor overall survival of ccRCC patients. Function assays revealed that circPCNXL2 knockdown reduced RCC cells proliferation, invasion in vitro, and decreased tumor growth in vivo. In mechanism study, we showed that circPCNXL2 could be bind to miR-153 as a miRNA sponge to regulate ZEB2 expression in RCC progression. In addition, our data reported that the effects of circPCNXL2 inhibition on RCC cells proliferation and invasion could be abolished by miR-153 inhibitors. Altogether, we demonstrated that circPCNXL2 could regulate RCC cells proliferation and invasion by miR-153/ZEB2 axis, suggesting circPCNXL2 might serve as a potential therapeutic target for ccRCC treatment.
Introduction
Renal cell carcinoma (RCC) is one of the most common tumors originated from renal tubular epithelial cells with a high mortality rate worldwides [Citation1,Citation2]. Clear cell renal cell carcinoma (ccRCC) is a major histologic subtype of RCC (approximately 70–80%) [Citation3]. Surgical resection is considered the primary treatment for RCC. However, the high rate of local or distant metastasis contributed to a dismal prognosis and a low 5-year survival rate of RCC patients [Citation4]. Therefore, improving the early diagnosis and exploring the new mechanism involved in RCC progression are very important.
With the rapid development of high-throughput sequencing technology, many non-coding RNAs (ncRNAs), previously regarded as junk molecules, have been identified and demonstrated to play vital roles in multiple physiological and pathological processes [Citation5,Citation6]. Circular RNAs (circRNAs) represent a new class of ncRNAs, which are characterized by high stability, covalently closed continuous loop, without 5’ to 3’ polarity and polyadenylated tail [Citation7]. Recently, accumulating evidence showed that circRNAs play important roles in tumor progression such as invasion, metastasis, and drug resistance [Citation8,Citation9]. For example, Zong et al showed that the expression of circRNA_102231was upregulated and associated with advanced TNM stage, lymph node metastasis, and poor overall survival of lung cancer patients [Citation10]. Li et al revealed that circRBMS3 promoted gastric cancer proliferation and invasion ability by regulating miR-153-SNAI1 axis [Citation11]. Zhu et al revealed that circPVT1 could contribute to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1 [Citation12]. These studies indicated that circRNAs might act as potential therapeutic targets for cancer treatment.
In the present study, we discovered a significantly up-regulated circRNA hsa_circRNA_406752 (circ-PCNXL2) utilizing human circRNA microarray analysis and explored the role of circPCNXL2 in RCC tissues and cells, providing a comprehensive landscape of circPCNXL2 in RCC progression.
Material and methods
Patients and specimens
A total of 63 pairs of ccRCC tissues and adjacent non-tumor tissues were derived from The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology between 2012 and 2013. None of them received any preoperative treatments prior to surgery. All ccRCC tissues were confirmed by two senior pathologists. All tissues were stored at liquid nitrogen until use. All patients were informed and consent was obtained. Our study was approved by the Ethics Committee of Henan University of Science and Technology.
Microarray analysis
Microarray analysis was performed by Arraystar Human circRNA Array V2, Fresh ccRCC tumor samples and matched non-tumor samples from 3 ccRCC patients were selected for microarray analysis. Total RNA from each sample was quantified using the NanoDrop ND-1000. The sample preparation and microarray hybridization were performed based on the Arraystar’s standard protocols [Citation13]. Agilent Feature Extraction software was used to analyze acquired array images. Quantile normalization and subsequent data processing was performed using the R software limma package. Differentially expressed circRNAs were identified through Fold Change filtering. Hierarchical Clustering was performed to show the distinguishable circRNAs expression pattern among samples.
Cell culture and transfection
Human RCC cell lines (A498, 786-O, ACHN, and Caki-1) and immortalized normal renal epithelial cell line HK-2 were used in this study. All cell lines were purchased from American Type Culture Collection (ATCC, Manassas, USA). All cells were cultured in Dulbecco’s Modified Essential Medium (DMEM, Gibco, Carlsbad, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, CA, USA) and antibiotics (1% penicillin–streptomycin) in a humidified incubator with 5% CO2 at 37°C.
Si-circPCNXL2, miR-153 mimics (miR-153), miR-153 inhibitors (anti-miR-153) and their respective controls were purchased from Gene-Pharma (Shanghai, China). Transfection were performed using the Lipofectamine 2000 reagents (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
RNA extraction and quantitative real time-pcr (qRT-PCR)
Total RNA was obtained from tissues and cell lines by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as previously reported [Citation14]. The quality of RNA was determined by using Nanodrop 2000 spectrophotometry (Thermo Scientific, USA). 500 ng of total RNA was reversely transcribed to cDNA using the PrimeScript RT reagent kit (Takara, Dalian, China). The expression analysis was performed using the SYBR Premix Ex Taq (Takara, Dalian, China) on LightCycler®480 system (Roche, Switzerland) according to the manufacturer’s instructions. GAPDH was employed as an internal reference gene, and relative expression fold changes were calculated using the 2−ΔΔCt method.
CCK-8 assay
Cell counting kit-8 (CCK-8) assay (Dojindo, Japan) was used to explored cell viability. Cells were added to a 96-well plate (3000 cells/well) and cultured for 24 h, 48 h, 72 h, and 96 h. Each group consisted of 6 wells and contained blank control group. After adding CCK-8 reagent for 1.5 h, the absorbance value of each well was measured by micro-plate reader instrument at 450 nm wavelength.
Colony formation assay
Transfected cells were seeded in a 6-well plate (1000 cells/well) and incubated for 12 days. We treated colonies with 0.1% crystal violet at room temperature for 15 min, and we counted and photographed cell colonies containing more than 50 cells.
Cell cycle analysis
The cells were harvested 48 h post‐transfection for cell‐cycle analysis. Cell suspension was obtained by digestion of each group, washed twice with PBS, and fixed overnight at 4 °C in 70% ethanol. Then, cells were treated with RNase A PI (Sigma, St Louis, MO, USA) for 20 min at room temperature. Cell cycle was determined with FACS flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Transwell invasion assay
Transfected cells were plated at 5 × 104 cells per well in 24-well plates with Matrigel (BD Biosciences, New York, NY, USA), while 600 µl medium containing 20% FBS were supplied at the lower chamber as chemo-attractant. The chambers were kept at 37 °C for 48 h. Afterwards, cells that invaded from the upper chamber to the lower chamber were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 30 min. Then stained cells were photographed and calculated in five random fields under an inverted microscope [Citation15].
In vivo assay
BALB/c nude mice (4 weeks old) were used in this study. Lentiviral vectors for circPCNXL2 (sh-circPCNXL2) and normal control (sh-NC) were separately constructed by GenePharm company. 786-O cells stably depleting circPCNXL2 were injected subcutaneously into the left of the armpit of each mouse (n = 3 for each group). The tumor volumes were calculated every 7 days in mice as volume = length × (width/2)2. Mice were sacrificed 42 days after injection and tumor weights were measured. The animal study was approved by the committee on animal experimentation of the Henan University of Science and Technology.
Luciferase reporter assay
Luciferase reporter vector with the full length of the 3′-UTR of ZEB2 or circPCNXL2 were constructed. Then we generated the mutant luciferase reporter vectors with QIAGEN XL-site directed Mutagenesis Kit (QIAGEN, California, USA). Cells were seeded into 96-well plates and co-transfected with luciferase reporter vector and miR-153 mimics or miR-NC using the Lipofectamine 2000 transfection reagent. After 48 h of incubation, the firefly and Renilla luciferase activities were quantified with a dual-luciferase reporter assay (Promega, Madison, WI, USA).
RNA immunoprecipitation (RIP)
RIP assay was carried out using the EZ-Magna RIP kit (Millipore) according to the protocol provided by the manufacturer [Citation16]. Briefly, whole-cell lysate was incubated with RIP buffer containing magnetic beads conjugated to a human anti-Ago2 antibody, or negative control normal mouse IgG. Samples lysis were all treated with Proteinase K, and the RNA immunoprecipitations were performed qRT-PCR assay to identify the presence in binding target antibody with respective primers.
RNA pull-down analysis
RNA pull-down analysis was performed as previously described [Citation17]. Briefly, the RIP lysates from cells were incubated with biotin (Bio)-labeled oligonucleotide probes against circPCNXL2 for 2 h at 25 °C. CircPCNXL2/miRNA complexes were captured with Streptavidin-coupled Dynabeads (Invitrogen). The complexes were incubated with RIP wash buffer (Millipore) containing proteinase K for 1 h at 25 °C. circPCNXL2 and miR-153 in the pull-down were determined using qRT-PCR analysis.
Western blot
Total protein was extracted from cells using RIPA lysis buffer (Beyotime, Shanghai, China). The lysate protein was separated by 10% SDS-PAGE and transferred onto PVDF membranes (Millipore, Bedford, MA, USA). Then, the membranes were incubated with primary antibody at 4°C overnight, followed by reaction with corresponding secondary antibody. GAPDH antibody was used as an internal control. ECL regent (Thermo Fisher Scientific, Waltham, MA, USA) was applied to develop the proteins.
Statistical analysis
All statistical analyses were carried out using SPSS 17.0 statistical software package and GraphPad Prism 7.0. All data were obtained from three independent experiments and expressed as mean ± standard deviation (SD). Student’s t-test (two-tailed) and one-way ANOVA were used compare two or more groups for statistical analysis, respectively. Kaplan-Meier analysis with log-rank tests were used for overall survive analysis. P values of < 0.05 were considered significant.
Results
CircPCNXL2 is upregulated and associated with ccRCC progression
A total of 3 ccRCC tissues and their adjacent non-tumor tissues were collected and screened for dysregulated circRNA using human circRNA microarray. Hierarchical clustering showed the top 10 upregulated and downregulated circRNAs in ccRCC tissues ()). QRT-PCR was used to verify the expression of the top 4 upregulated circRNAs in 15 paired ccRCC tissues ()). The hsa_circRNA_406752 was the top upregulated circRNA in ccRCC tissues. According to the human reference genome, we further termed hsa_circRNA_406752 (located at chr1:233152666-233397067, is derived from gene PCNXL2, which is located on chromosome 1q42.2.) as “circPCNXL2”.
Figure 1. CircPCNXL2 is upregulated in clear cell renal cell carcinoma (ccRCC). (a) A heat map showed the top 10 up- and downregulated circRNAs in ccRCC tissues (T) and adjacent non-tumor tissues (NT). (b) QRT-PCR determined 4 indicated circRNAs expression listed in (a) from 15 ccRCC tissues and adjacent non-tumor tissues (NT). (c) Relative circPCNXL2 expression in ccRCC tissues and adjacent non-tumor tissues was determined by qRT-PCR. (d) High circPCNXL2 expression was associated with poor overall survival of ccRCC patients. (e) Relative circPCNXL2 expression in RCC cell lines (A498, 786-O, ACHN, and Caki-1) and HK-2 cells was determined by qRT-PCR. *P < 0.05.
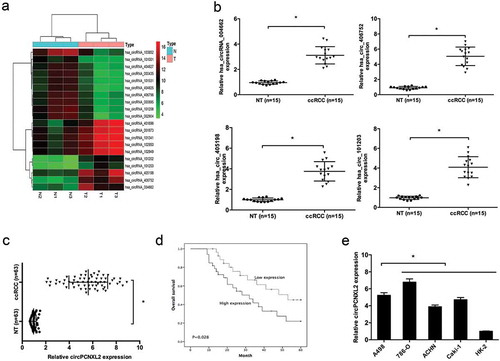
Next, we explored circPCNXL2 expression in 63 pairs of ccRCC tissues. QRT-PCR results showed that circPCNXL2 expression was significantly upregulated in ccRCC tissues compared to adjacent non-tumor tissues (NT) (); P < 0.05). According to the median ratio of circPCNXL2 expression, patients were divided into high circPCNXL2 expression group and low circPCNXL2 expression group. Kaplan–Meier analysis and log-rank test showed that patients with high circPCNXL2 expression had a poor overall survival (); P < 0.05). In addition, qRT-PCR showed that circPCNXL2 expression was dramatically increased in RCC cell lines (A498, 786-O, ACHN, and Caki-1) compared to HK-2 cells, especially in A498 and 786-O cells (); P < 0.05).
Circpcnxl2 inhibition decreases RCC cells proliferation and invasion in vitro
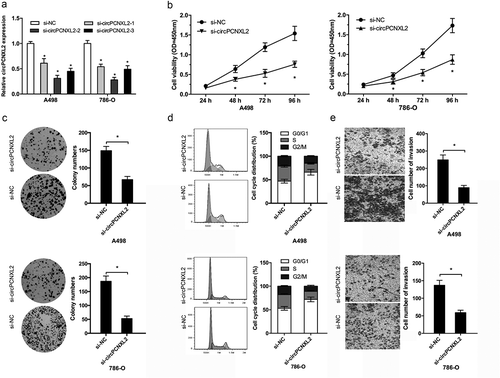
To explored the roles of circPCNXL2, siRNAs were used to knockdown circPCNXL2 expression in A498 and 786-O cells (); P < 0.05). CCK-8 assay showed that circPCNXL2 inhibition significantly reduced A498 and 786-O cells proliferation ability (); P < 0.05). Knockdown of circPCNXL2 inhibited A498 and 786-O cells colony formation ability (); P < 0.05). Flow cytometry analysis showed that circPCNXL2 reduction arrested A498 and 786-O cells in G0/G1 phase (); P < 0.05). Moreover, transwell assay revealed that circPCNXL2 suppression significantly decreased A498 and 786-O cells invasion ability (); P < 0.05). These findings indicated that circPCNXL2 might be closely associated with the proliferation and invasion of RCC cells.
Figure 2. CircPCNXL2 inhibition reduces RCC cells proliferation and invasion in vitro. (a) QRT-PCR analysis for circPCNXL2 expression in A498 and 786-O cells transfected with si-circPCNXL2. (b, c) CircPCNXL2 inhibition decreased RCC cells colony formation ability and cell proliferation. (d) CircPCNXL2 suppression arrested RCC cells in G0/G1 phase. (e) CircPCNXL2 inhibition reduced the invasion ability of RCC cells. *P < 0.05.
Circpcnxl2 inhibition reduces tumor growth in vivo
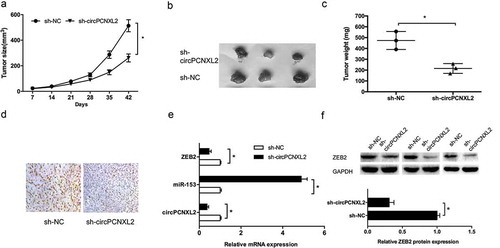
To evaluate effects of circPCNXL2 on tumorigenesis in vivo, we injected 786-O cells expressing either sh-circPCNXL2 or empty vector subcutaneously into nude mice for xenograft implantation. Results showed that circPCNXL2 inhibition significantly decreased tumor growth in vivo (Figure 3(a–); P < 0.05). Moreover, sh-circPCNXL2 expressing xenograft tumors displayed decreased Ki67 expression (); P < 0.05). In addition, we explored circPCNXL2, miR-153 and ZEB2 expression in xenograft tumors, results showed that circPCNXL2 and ZEB2 expression was significantly decreased, while miR-153 expression was increased in sh-circPCNXL2 group (,); P < 0.05).
Figure 3. CircPCNXL2 inhibition reduces tumor growth in vivo. (a) Tumor volume was determined by caliper on the indicated days. (b) Photographs of tumors excised 42 days of injection. (c) Tumors weight was detected after 42 days of injection. (d) IHC of Ki-67 expression in xenograft tumors after 42 days of injection. (e) The expression of circPCNXL2, miR-153 and ZEB2 in xenograft tumors was detected by qRT-PCR. (f) The expression of ZEB2 in xenograft tumors was detected by Western blot. *P < 0.05.
Circpcnxl2 binds to mir-153 in RCC cells
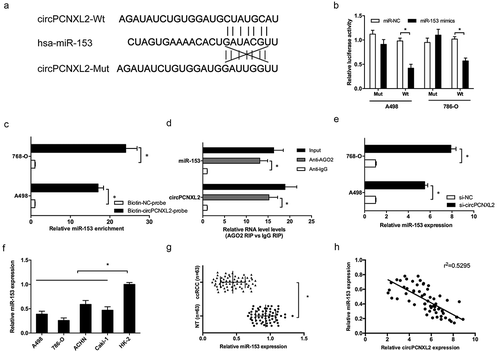
Recently, increasing evidence showed that circRNA may work as competing endogenous RNA to interact with miRNAs in pathological [Citation18]. In the present study, we predicted the potential interactive miRNAs of circPCNXL2 used miRBase and circinteractome. Results showed that circPCNXL2 possesses a potential binding site for miR-153 ()). Dual-luciferase reporter assay showed that miR-153 mimics significantly inhibited the luciferase activity of circPCNXL2-Wt reporter (); P < 0.05). RNA pull‐down assay using biotin‐labeled circPCNXL2 probes revealed that circPCNXL2 precipitated miR‐153 in RCC cells (); P < 0.05). Next, we performed RIP experiments using anti-Ago2 in 786-O cells extracts. Results showed that circPCNXL2 and miR-153 were enriched in Ago2-containing miRNPs compared to anti-IgG immunoprecipitates (); P < 0.05). QRT-PCR revealed that circPCNXL2 inhibition increased miR-153 expression (); P < 0.05). In addition, qRT-PCR showed that miR-153 expression was significantly decreased in RCC cells and ccRCC tissues (,); P < 0.05). Correlation analysis revealed that miR-153 expression was negatively correlated with circPCNXL2 expression in ccRCC tissues (); P < 0.05).
Figure 4. CircPCNXL2 binds to miR-153 in RCC cells. (a) The estimated binding sites of circPCNXL2 and miR-153. (b) MiR-153 mimics reduced the luciferase activity of circPCNXL2-Wt reporter. (c) RNA pull-down assay showed that circPCNXL2 interacted with miR-153 directly. (d) RIP assay showed that circPCNXL2 and miR-153 were enriched in Ago2-containing miRNPs. (e) CircPCNXL2 inhibition increased miR-153 expression in RCC cells. (f, g) MiR-153 expression was reduced in RCC cell lines and ccRCC tissues. (h) CircPCNXL2 expression was negatively correlated with miR-153 expression in ccRCC tissues. *P < 0.05.
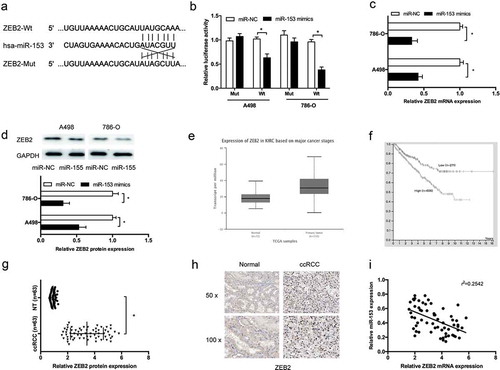
ZEB2 is a target of mir-153
Next, ZEB2 was identified as a potential target of miR-153 by Targetscan ()). To confirm the speculation, dual-luciferase reporter assay revealed that miR-153 mimics significantly reduced the luciferase activity of ZEB2-Wt 3’-UTR reporter (); P < 0.05). QRT-PCR and Western blot assays showed that miR-153 mimics significantly decreased ZEB2 expression in RCC cells (,); P < 0.05). TCGA database showed that ZEB2 expression was significantly increased and correlated with poor overall survival of ccRCC patients (,); P < 0.05). To confirmed it, QRT-PCR and IHC were used to determine ZEB2 expression in ccRCC tissues. Result showed that ZEB2 expression was significantly increased in ccRCC tissues (,); P < 0.05). In addition, correlation analysis showed that miR-153 expression was inversely correlated with ZEB2 mRNA expression in ccRCC tissues (); P < 0.05).
Figure 5. ZEB2 is a target of miR-153. (a) The estimated binding sites of ZEB2 and miR-153. (b) MiR-153 mimics reduced the luciferase activity of ZEB2-Wt reporter. (c, d) MiR-153 mimics decreased ZEB2 expression both in mRNA and protein levels. (e) TCGA database showed that ZEB2 was highly expressed in ccRCC tissues. (f) High ZEB2 expression contributed to a significant poor overall survival of ccRCC in TCGA database. (g, h) Relative ZEB2 expression was increased in ccRCC tissues determined by qRT-PCR and IHC. (i) MiR-153 expression was negatively correlated with ZEB2 expression in ccRCC tissues. *P < 0.05.
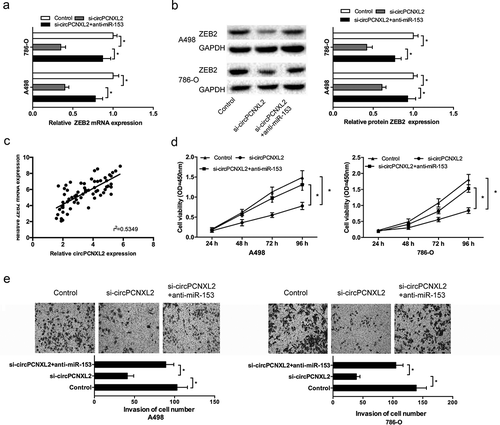
Circpcnxl2 inhibition reduced RCC cells progression by mir-153/zeb2 axis
Next, we explored whether circPCNXL2 promoted RCC progression by modulating miR-153/ZEB2 axis. We found that circPCNXL2 knockdown significantly decreased ZEB2 expression both in mRNA and protein levels in A498 and 786-O cells, and miR-153 inhibitors (anti-miR-153) at least partly reversed the effects (Figure 6(a,); P < 0.05). Correlation analysis revealed that circPCNXL2 expression was positively correlated with ZEB2 mRNA expression in ccRCC tissues (); P < 0.05). In addition, function assays showed that miR-153 inhibitors could reverse the effects of circPCNXL2 inhibition on RCC cells proliferation and invasion (,); P < 0.05).
Figure 6. CircPCNXL2 inhibition reduced RCC progression by miR-153/ZEB2 axis. (a, b) CircPCNXL2 suppression decreased ZEB2 expression both in mRNA and protein levels, which could be reversed by miR-153 inhibitors (anti-miR-153). (c) CircPCNXL2 expression was positively correlated with ZEB2 mRNA expression in ccRCC tissues. (d, e) CCK-8 and transwell invasion assays revealed that miR-153 inhibitors could abolish the effects of circPCNXL2 inhibition on RCC cells progression. *P < 0.05.
Discussion
CircRNA is a covalently closed, single-stranded transcripts and identified as a naturally occurring ncRNA family [Citation19]. Increasing evidence revealed that dysregulation of circRNAs are associated with the malignant behaviors in urinary cancers. For example, Zeng et al revealed that circ-VANGL1 acted as a ceRNA contributed to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway [Citation20]. Dai et al showed that circular RNA MYLK promoted prostate cancer progression through modulating miR-29a expression [Citation21]. Wang et al found that androgen receptor (AR) promoted ccRCC cells migration and invasion via circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 axis [Citation22]. Current studies indicated that circRNAs play an important role in urinary tumors progression. However, the role of circRNAs in RCC is still unclear. In the present study, we found that circPCNXL2 was significantly upregulated and positively correlated with poor overall survival of ccRCC patients. Function assays showed that circPCNXL2 knockdown could decrease RCC cells proliferation, invasion in vitro and reduce tumor growth in vivo. Those data indicated that circPCNXL2 could serve as an oncogenic circRNA in RCC progression.
Recently, increasing studies have shown that circRNAs play critical roles in regulating gene expression by sequestering the miRNAs [Citation23]. In the present study, we investigated whether circPCNXL2 could regulate miRNA expression in RCC progression. Based on miRBase and circinteractome software, we verified that circPCNXL2 possessed a potential binding site for miR-153. Through luciferase reporter, RNA pull-down, and RIP assays, we further confirmed their direct interaction. Recently, previous studies showed that miR-153 could act as a tumor suppressor in a variety of cancers. For example, Zhang et al showed that miR-153 suppressed human laryngeal squamous cell carcinoma migration and invasion by targeting the SNAI1 [Citation24]. Ghasemi et al revealed that miR-153 acted as a tumor suppressor in glioblastoma multiforme and was downregulated by DNA methylation [Citation25]. In the present study, we showed that miR-153 was markedly decreased in ccRCC tissues and cell lines. And there was a negative correlation between miR-153 and circPCNXL2 in ccRCC tissues. Those data indicated that circPCNXL2 might serve as a sponge of miR-153 in RCC progression.
Zinc finger E-box binding homeobox 2 (ZEB2), a member of the zinc finger family, functions as a transcriptional repressor of E-cadherin, which play critical roles in various cellular processes, including cell proliferation, differentiation, and migration [Citation26]. Kahlert et al revealed that overexpression of ZEB2 at the invasion front correlated with tumor progression and predicted cancer-specific survival in primary colorectal cancer [Citation27]. Xiao et al found that lncRNA MALAT1 functioned as a ceRNA to regulate ZEB2 expression by sponging miR-200s in ccRCC [Citation28]. Although its oncogenic roles are acknowledged in some cancers, its role in ccRCC remains unclear. In the present study, we showed that ZEB2 is a direct target of miR1-153. Furthermore, we showed that ZEB2 could be reduced by si-circPCNXL2, and miR-153 inhibitors could abolish the effects. Through rescue assay, we demonstrated that miR-153 inhibitors could reverse the effects of circPCNXL2 inhibition on RCC cells progression.
In conclusion, our results indicated that circPCNXL2 could serve as an oncogenic circRNA to promote RCC progression by modulating the miR-153/ZEB2 axis. Thus, our finding suggested that circPCNXL2 could serve as a potential therapeutic target for RCC treatment.
Acknowledgments
None.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353(23):2477–2490.
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
- Ljungberg B, Campbell SC, Cho HY, et al. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60(4):615–621.
- Znaor A, Lortet-Tieulent J, Laversanne M, et al. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol. 2015;67(3):519–530.
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874.
- R J T, K C P, Mercer TR, et al. Non coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461.
- Chen L-L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211.
- Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. 2018;102:639–644.
- Li G, Xue M, Yang F, et al. CircRBMS3 promotes gastric cancer tumorigenesis by regulating miR 153–SNAI1 axis. J Cell Physiol. 2019;234(3):3020–3028.
- Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14(3):321.
- Chen B, Wei W, Huang X, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8(14):4003–4015.
- Zhao F, Han Y, Liu Z, et al. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep. 2018;38:BSR20180570 (Epub Ahead of Print).
- Shahid M, Kim M, Yeon A, et al. Quantitative proteomic analysis reveals caffeine perturbed proteomic profiles in normal bladder epithelial cells. Proteomics. 2018;18(20):1800190.
- Hu C, Wang Y, Li A, et al. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR 545/EIF3C axis. J Cell Physiol. 2018 (Epub Ahead of Print).
- Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151.
- Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51.
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA 9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164.
- Zeng Z, Zhou W, Duan L, et al. Circular RNA circ VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR 605 3p/VANGL1 pathway. J Cell Physiol. 2018(Epub Ahead of Print).
- Dai Y, Li D, Chen X, et al. Circular RNA Myosin Light Chain Kinase (MYLK) promotes prostate cancer progression through modulating Mir-29a expression. Med Sci Monit. 2018;24:3462–3471.
- Wang K, Sun Y, Tao W, et al. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12.
- Ebbesen KK, Kjems J. microRNA sponging by a new hearty circRNA. Non-Cod RNA Investgat. 2017;1(3):6–7.
- Zhang B, Fu T, Zhang L. MicroRNA-153 suppresses human laryngeal squamous cell carcinoma migration and invasion by targeting the SNAI1 gene. Oncol Lett. 2018;16(4):5075–5083.
- Ghasemi A, Fallah S, Ansari M. MiR-153 as a tumor suppressor in glioblastoma multiforme is downregulated by DNA methylation. Clin Lab. 2016;62(4):573–580.
- Nam EH, Lee Y, Park YK, et al. ZEB2 upregulates integrin α5 expression through cooperation with Sp1 to induce invasion during epithelial–mesenchymal transition of human cancer cells. Carcinogenesis. 2012;33(3):563–571.
- Kahlert C, Lahes S, Radhakrishnan P, et al. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clin Cancer Res. 2011;17(24):7654–7663.
- Xiao H, Tang K, Liu P, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6(35):38005.
