ABSTRACT
Meiosis is the basis for sexual reproduction and is marked by the sequential reduction of chromosome number during successive cell cycles, resulting in four haploid gametes. A central component of the meiotic program is the formation and repair of programmed double strand breaks. Recombination–driven repair of these meiotic breaks differs from recombination during mitosis in that meiotic breaks are preferentially repaired using the homologous chromosomes in a process known as homolog bias. Homolog bias allows for physical interactions between homologous chromosomes that are required for proper chromosome segregation, and the formation of crossover products ensuring genetic diversity in progeny. An important aspect of meiosis in the differential regulation of the two eukaryotic RecA homologs, Rad51 and Dmc1. In this review we will discuss the relationship between biological programs designed to regulate recombinase function.
KEYWORDS:
Introduction
Meiosis is a complex biological program that is required for sexual reproduction and is characterized by two successive chromosome divisions (Meiosis I and Meiosis II), each step resulting in reduced chromosome copy number (Figure 1(a)). In meiosis I homologous chromosomes are paired and separated, meiosis I is followed by meiosis II in which sister chromatids are separated. The net result of both meiotic divisions is the formation of haploid gametes that will become an organisms next generation. Meiosis I can be further separated into leptotene, zygotene during which homologous chromosomes come into alignment, pachytene, during which DNA crossovers form between homologous chromosomes, diplotene in which homologous chromosomes begin to separate, and diakinesis, in which crossovers are resolved and homologous chromosomes separated ()) [Citation1]. These stages of the meiotic program are dependent on the formation and repair of programmed DNA double strand breaks (DSBs). S. cerevisiae has long served as a paradigm for studying meiosis, in part due to its ability to cycle through haploid and diploid states, the ability to recover all four gametes, and the fact that many of the steps involved in meiosis are broadly conserved [Citation2].
Figure 1. Chromosomal changes that take place during meiosis. Diagram illustrating the major steps in meiosis. Meiosis proceeds through a single cycle of DNA replication followed by two cycles of cell division, resulting in a reduction in chromosome copy number to yield haploid spores or gametes. We refer the reader to several recent reviews for more in–depth descriptions of the mechanisms underlying meiosis [Citation1,Citation3,Citation33].
![Figure 1. Chromosomal changes that take place during meiosis. Diagram illustrating the major steps in meiosis. Meiosis proceeds through a single cycle of DNA replication followed by two cycles of cell division, resulting in a reduction in chromosome copy number to yield haploid spores or gametes. We refer the reader to several recent reviews for more in–depth descriptions of the mechanisms underlying meiosis [Citation1,Citation3,Citation33].](/cms/asset/0c87ea6b-cd0d-4b67-b916-785e60ed142f/kccy_a_1553355_f0001_oc.jpg)
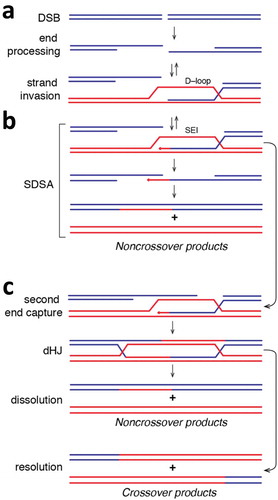
A key regulatory step during meiosis is the formation of programmed DSBs, which are repaired by homologous recombination (HR). HR is a universally conserved DNA repair pathway that has been confiscated in eukaryotes as a means to insure proper segregation of homologous chromosomes during meiosis I [Citation1,Citation3]. Meiotic HR is initiated by the universally conserved Spo11 endonuclease [Citation4], which uses a tyrosine amino acid residue to cleave dsDNA [Citation4,Citation5]. The DNA is then resected by Mre11–Rad50–Xrs2 (MRX), Exo1, and Sae2, which removes Spo11 and also generates long 3ʹ single strand DNA (ssDNA) overhangs [Citation6,Citation7]. The heterotrimeric ssDNA–binding protein RPA binds to these overhangs to protect the ssDNA and prevent formation of secondary structure [Citation8–Citation10]. The recombinases Rad51 and Dmc1 are then loaded onto the ssDNA [Citation11,Citation12] through a poorly understood mechanism involving the mediator proteins Rad52 [Citation13] and Mei5–Sae3 [Citation14,Citation15] to establish the presynaptic complex. A number of accessory proteins also bind to the presynaptic complex and stimulate its activities, including but not limited to Rad54, Rdh54, Hop2–Mnd1, and Hed1 [Citation1,Citation16,Citation17]. The presynaptic complex then searches for a homologous target and catalyzes strand invasion wherein the 3ʹ ssDNA overhang is paired with a complementary strand from the homologous chromosome to yield a D–loop intermediate, which is thought to be unstable in vivo (Figure 2) [Citation18,Citation19]. D–loops can be extended by a DNA polymerase, yielding a more stable single end invasion intermediate (SEI) () [Citation20–Citation22]. SEIs can then be disrupted, allowing for repair through the synthesis dependent strand annealing pathway (SDSA) to yield non–crossover recombination products. Alternatively, second end capture allows for conversion of the SEIs into double Holliday junctions (dHJs), which can allow for the formation of crossover recombination products () [Citation23]. Chiasmata form at a defined number of crossover sites, creating a physical linkage between homologous chromosomes [Citation24–Citation27]. In yeast, the total number of DSBs that form are roughly 10–fold greater than the number of crossovers, indicating most DSBs are still repaired by non–crossover pathways [Citation24,Citation28]. This low number of crossovers is regulated by crossover interference, a poorly understood phenomenon that limits the number of connecting points between chromosomes [Citation1,Citation24–Citation27].
Figure 2. Simplified overview of homologous recombination. (a) Diagram depicting the formation of a DSB followed by end processing of the break to yield long 3’ssDNA overhangs. The ssDNA can be paired with a homologous dsDNA (shown in red) to yield a metastable D–loop intermediate. The 3ʹend of the invading strand of the D–loop can then be used to prime DNA synthesis, which yields a more stable single end invasion product (SEI). (b) The SEI can then channeled through the synthesis–dependent strand annealing pathway. In this case, the SEI product is displaced from the homologous template and can repair with the second processed DNA end. Additional DNA synthesis followed by strand ligation yields noncrossover repair products. (c) Alternatively, the second end of the processed DNA can anneal to the homologous template, which can result in the formation of an interlinked double Holliday junction (dHJ) after DNA synthesis and strand ligation. The dHJ can be dissolved to yield noncrossover products, or it can undergo resolution through endonucleolytic cleavage, which can yield either noncrossover products (not depicted) or crossover products.
Although they share many central features, there are a number of things that distinguish meiotic and mitotic recombination, including differences in the Rad51/RecA family recombinases that catalyze the underlying DNA transactions, the participating regulatory protein co–factors, the chromosome structures, changes in the relative ratios of crossover to non–crossover products, and differences in the identity of the templates that are used to guide DSB repair [Citation1,Citation3,Citation29,Citation30]. In this review we briefly highlight the biochemical mechanisms that underlie differences between mitotic and meiotic HR, with emphasis on studies involving proteins from S. cerevisiae.
Biochemical characteristics of Rad51 and Dmc1
The vast majority of eukaryotes require two recombinases: Rad51, which is expressed during both mitosis and meiosis, and Dmc1 is only expressed during meiosis [Citation11,Citation31] . Dmc1 and Rad51 emerged from a gene duplication event during the early evolutionary history of eukaryotes [Citation32], and the recombinases retain ~46% amino acid identity and closely related structural and biochemical properties [Citation18,Citation33,Citation34]. Such strong similarities beg the question why most eukaryotes require a specialized recombinase for meiosis.
There are some biochemical differences between Rad51 and Dmc1 that may be related to their roles in mitotic and meiotic recombination, respectively. For instance, one clearly important difference between Rad51 and Dmc1 is that they interact with distinct subsets of recombination factors [Citation1,Citation3,Citation30]. For example, the meiosis–specific protein Hed1 binds specifically to Rad51 and down–regulates its strand exchange activity (see below). Similarly, Dmc1 interacts specifically with the mediator complex Mei5–Sae3, which stimulates Dmc1 filament assembly [Citation35], and the accessory factor Hop2–Mnd1, which promotes dsDNA capture by the presynaptic complex [Citation36–Citation39]. In addition, there is biochemical evidence suggesting that the recombination accessory proteins Rad54 and Rdh54 interact preferentially with Rad51 and Dmc1, respectively [Citation40,Citation41]. Biochemical studies have also shown that Dmc1 requires divalent calcium ions for strand exchange activity and filament stability in vitro whereas Rad51 does not [Citation36,Citation42,Citation43]It remains to be seen whether calcium reflects a physiological requirement of Dmc1 or whether Ca 2+ may somehow mimic the stimulatory effects of an as yet unknown protein cofactor.
Interestingly, biophysical studies have shown that Rad51–ssDNA filaments can bind to dsDNA fragments containing short tracts of sequence microhomology, but the introduction of a single nucleotide mismatch causes a reduction in binding lifetime commensurate with the loss of one base triplet pairing interaction. In contrast, Dmc1–ssDNA can tolerate base triplets bearing single, double, or triple mismatches and even abasic sites with no change in the binding lifetimes of the heteroduplex DNA intermediates [Citation44,Citation45]. These findings suggest that Dmc1 can stabilize heteroduplex DNA joints containing mismatched base triplets, whereas Rad51 cannot. We have previously hypothesized that the ability of Dmc1 to stabilize imperfectly paired recombination intermediates might make Dmc1 better equipped to stabilize mismatches present in heteroduplex recombination intermediates formed between homologous chromosomes of different parental origins [Citation46–Citation48]. However, the molecular basis for these biophysical differences, and their biological implications remained unexplored.
In addition to interacting with distinct protein co–factors, Rad51 and Dmc1 also interact differently with one another. Indeed, early microscopy studies suggested that Rad51 and Dmc1 formed foci that were close to one other, but did not precisely overlap, suggesting the possibility that they may be making separate filaments on alternate ends of the DSBs ()) [Citation49]. However, recent advances in super–resolution microscopy revealed that Rad51 and Dmc1 are co–localized to both 3ʹ ssDNA overhangs present on each DSB end, resulting in mixed recombinase filaments organized in a side–by–side configuration ()) [Citation12]. Surprisingly, this mixed recombinase side–by–side organization can be reconstituted in vitro without the for any recombination mediators, indicating that all of the information necessary for segregation of the homotypic filaments was encoded within the recombinases themselves [Citation43]. One implication of this type of highly segregated filament organization is that the Rad51 – and Dmc1–specific binding factors that associate with the meiotic presynaptic are themselves going to be spatially segregated, reflecting the underlying separation of Rad51 and Dmc1 into homotypic filaments ()). How the spatial organization of all these proteins impacts meiotic recombination remains to be determined. Interestingly, Dmc1 also appears to stabilize the adjacent Rad51 filaments to dissociation upon depletion of ATP, suggesting that cross–talk between these two recombinases may affect their biochemical properties [Citation43]. One possible implication of this finding is that protein factors bound to one of the two recombinases may influence the properties of the other recombinase through filament–mediated allosteric communication.
Figure 3. Spatial organization of protein components within the meiotic presynaptic complex. (a) Diagram illustrating the “alternate end” and “mixed filament” models describing the organization of the Rad51 and Dmc1 recombinases at the processed DSBs formed through the endonuclease active of Spo11 during meiosis. Each model invokes the formation of homotypic Rad51 and Dmc1 filaments (as depicted) and current research favors the “mixed filament” model [Citation11,Citation43]. (b) Cartoon model depicting the hypothetical spatial organization of recombination accessory factors based upon their binding interactions with the Rad51 or Dmc1 and the mixed filament model for meiotic presynaptic complex. The locations of some of the factors remains unknown, and a more detailed understanding of the protein binding mechanisms and protein distributions remains an active area of investigation. See main text for additional details.
![Figure 3. Spatial organization of protein components within the meiotic presynaptic complex. (a) Diagram illustrating the “alternate end” and “mixed filament” models describing the organization of the Rad51 and Dmc1 recombinases at the processed DSBs formed through the endonuclease active of Spo11 during meiosis. Each model invokes the formation of homotypic Rad51 and Dmc1 filaments (as depicted) and current research favors the “mixed filament” model [Citation11,Citation43]. (b) Cartoon model depicting the hypothetical spatial organization of recombination accessory factors based upon their binding interactions with the Rad51 or Dmc1 and the mixed filament model for meiotic presynaptic complex. The locations of some of the factors remains unknown, and a more detailed understanding of the protein binding mechanisms and protein distributions remains an active area of investigation. See main text for additional details.](/cms/asset/6b5a26b0-827e-446a-864a-e2b81100e0f1/kccy_a_1553355_f0003_oc.jpg)
Although there are some biochemical differences between Rad51 and Dmc1, as yet none of these differences definitely explains the evolutionary pressure that allowed for the initial emergence of Dmc1 as a necessary factor for the meiotic program. Some speculative possibilities are that Dmc1 evolved to interact specifically with another protein, which provided some specific benefit for early iterations of the meiotic program. Alternatively, Dmc1 may have lower recombination fidelity compared to Rad51, which may have been beneficial for promoting recombination between imperfectly matched sequences, thus generating genetic diversity, whereas a higher fidelity Rad51 may have been better for maintaining genome stability during mitosis. Finally, the ability of Rad51 and Dmc1 to self–segregate may have been an early evolutionary step necessary to allow for further specialization of the separate Rad51 and Dmc1 lineages within the Rad51/RecA family.
Down regulation of Rad51 strand exchange activity during meiosis
Dmc1 catalyzes strand exchange during meiosis, whereas the strand exchange activity of Rad51 is down regulated during meiosis [Citation50,Citation51]. To understand the mechanism of Rad51 down regulation we must first consider the relationship between Rad51 and Rad54. Rad54 is an ATP–dependent Sf2 dsDNA translocase that is an essential Rad51 co–factor during mitosis [reviewed in refs. [Citation16,Citation52]]. Indeed, deletion of the RAD54 gene is just as deleterious to cell survival as is deletion of RAD51 [Citation53–Citation56]. Exactly how Rad54 works in vivo remains uncertain, but a number of biochemical activities attributed to Rad54 may promote Rad51–dependent recombination. For instance, Rad54 may enhance the rate of the homology search [Citation57], Rad54 can alter the topology of duplex DNA to promote pairing interactions during the homology search and/or strand invasion [Citation58], Rad54 also remodels nucleosomes to ensure that Rad51 can gain access to dsDNA sequences otherwise embedded in chromatin [Citation59], and Rad54 removes Rad51 from the heteroduplex product of strand invasion to ensure that the 3ʹOH of the invading strand in available as a primer for DNA synthesis [Citation54,Citation60–Citation62].
Cells utilize of the interdependence of Rad54 and Rad51 as a means of selectively down regulating Rad51 during meiosis ()). In S. cerevisiae the down regulation of Rad51 occurs through two pathways involving (i) the meiosis–specific Rad51–binding protein Hed1 [Citation63,Citation64] and (ii) the meiosis–specific protein kinase Mek1 [Citation63–Citation67]. Hed1 was discovered as a high–copy suppressor of a cold sensitive allele of Red1, a component of the synaptonemal complex (SC; )) [Citation51]. Ectopic expression of Hed1 in mitotically growing cells inhibits Rad51–dependent repair of DNA damage [Citation68]. Biochemical studies revealed that Hed1 prevents the association of Rad54 with Rad51 through competitive inhibition [Citation50,Citation68,Citation69]. Single–molecule biophysical studies confirmed that Hed1 prevents the association of Rad54 with Rad51 filaments, while having no impact upon the ability of Rad54 to interact with Dmc1 filaments [Citation69]. These studies also demonstrated that the binding of either Hed1 or Rad54 was essentially irreversible on biologically relevant time scales, indicating that whichever protein bound to Rad51 first would likely define the functionality of the presynaptic complex [Citation69]. One possible implication of this result is that if Rad54 were to engage a meiotic presynaptic complex before Hed1 could bind, then that particular DSB might be preferentially repaired using the sister chromatid as a template, conversely, if Hed1 were to bind before Rad54, then these DSBs might be selectively repaired using the homologous chromosome as a template [Citation69]. Interestingly, there is no Hed1 homolog in higher eukaryotes, so it remains to be seen whether Rad51 strand exchange activity is down–regulated during meiosis in higher organisms.
Mek1 is a kinase that is essential for progression through meiosis [Citation65,Citation67,Citation70] and is considered to be a master regulator of the meiotic program [Citation70] . Mek1 phosphorylates a number of proteins, including Rad54 and Hed1. Mek1–dependent phosphorylation of Rad54 amino acid residue threonine 132, which is located in the N–terminal Rad51 – and ssDNA–binding domain of Rad54, reduces the ability of Rad54 to interact with Rad51–ssDNA filaments, thus reducing the strand exchange activity of Rad51 [Citation64]. Mek1 phosphorylates Hed1 on amino acid residue threonine 40, which stabilizes the Hed1 protein in meiotic cells, thus enhancing the ability of Hed1 to down-regulate Rad51 activity [Citation65]. In addition to acting as a co–factor for Rad51, Rad54 also interacts with Dmc1, but it is not yet known whether this interaction is similarly affected by Mek1–dependent phosphorylation. Mek1 also phosphorylates the Rad54 homolog Rdh54, which also binds to Rad51 and is partially redundant with Rad54. However, there are no phenotypes associated with Rdh54 mutations in the phosphorylated residues, suggesting that Mek1 phosphorylation of Rdh54 is less likely to play a role in regulating meiosis [Citation64]
Up regulation of Dmc1 during meiosis
There are number of mechanisms in place to help ensure the Dmc1 is the active recombinase that promotes strand invasion in meiosis. First, expression of the DMC1 gene is under control of a meiosis–specific promoter, allowing for rapid expression of high levels of Dmc1 upon entry into meiosis [Citation71]. Interestingly, introns are present in <5% of the genes in S. cerevisiae, so it is notable that several meiosis–specific genes, including Hop2, Mnd1, Sae3, Rec114, and Dmc1, all have introns. Thus, it is also possible that the expression of these meiosis–specific genes may also be regulated at the level of mRNA splicing [Citation72]. In addition to being regulated at the level of mRNA production, the activity of Dmc1 is directly affected through the actions of several meiosis–specific protein co–factors [Citation14,Citation36,Citation73]. For example, the protein complex Mei5–Sae3 is necessary for Dmc1 filament assembly [Citation74]. Although the molecular mechanisms remain poorly understood, Mae5–Sae3 appears to function in a manner similar to the Rad51–specific mediator protein Rad52, which is necessary to promote the assembly of Rad51 filaments in RPA–coated ssDNA in vivo [Citation14,Citation73]. In addition, S. cerevisiae Dmc1 strand exchange activity is greatly stimulated by the meiosis–specific protein complex Hop2–Mnd1 [Citation36,Citation37,Citation75,Citation76]. However, S. cerevisiae Hop2–Mnd1 does not interact with or stimulate the strand exchange activity of Rad51 [Citation33]. Structural studies reveal that Hop2–Mnd1 forms a long rod–like structure made up of three leucine zippers [Citation77] . The leucine zippers are capped on one end with two dsDNA–binding winged–helix domains, and the other end is capped with an α–helical bundle, which interacts with the Dmc1 filament [Citation77]. This bi-functional organization suggests a model in which one end of the Hop2–Mnd1 complex may interact with the Dmc1–ssDNA filament, while the other end binds to potential dsDNA targets, the effect of which might be to stimulate dsDNA capture by Dmc1 ()). Interestingly, the meiosis–specific simulation of Dmc1 by Hop2–Mnd1 is not conserved among higher organisms, for instance, in mice and humans HOP2–MND1 interacts with and stimulates both RAD51 and DMC1. Future work will be necessary to more fully understand the impact of Hop2–Mnd1 on yeast Dmc1, and to determine why yeast Hop2–Mnd1 does not interact with Rad51.
Crossover regulation by helicases
Crossovers are essential for the successful completion of meiosis [Citation1,Citation27,Citation78,Citation79], however, crossovers can also lead to potentially lethal gross chromosomal rearrangements [Citation80,Citation81]. Therefore, the pathways that lead to crossover recombination products are down regulated during mitotic growth, and recombination intermediates are instead channeled through the synthesis–dependent strand annealing pathway (SDSA), which leads to non–crossover recombination products () [Citation16,Citation82,Citation83]. In S. cerevisiae the helicases Srs2, Sgs1 and Mph1 have all been implicated as important negative regulators of crossovers during mitotic growth [Citation81,Citation84–Citation86]. Srs2 is a Sf1 family helicase that shares homology with the bacterial UvrD protein [Citation87,Citation88]. Srs2 dismantles Rad51–ssDNA filaments during the early stages of HR [Citation89–Citation93]. Mph1 the yeast homolog of human Sf2 helicase FANCM (Fancomi Anemia complementation group M) [Citation94,Citation95] that disrupts D–loops [Citation96]. Sgs1 is a RecQ helicase that is homologous to the human helicase BLM, which is mutated in a number of human diseases and cancer [Citation97]. The primary functions of Sgs1 appear to be in promoting DSB end resection during the earliest stages of HR [Citation7,Citation98–Citation100], and in catalyzing dissolution of double Holliday Junctions during the later stages of HR [Citation81,Citation101,Citation102], and Sgs1 and related RecQ helicases have also been implicated in D–loop disruption [Citation103]. Thus, Srs2, Mph1 and Sgs1 minimize crossover formation by disrupting HR intermediates that would otherwise led to crossovers, and their ability to disrupt these HR intermediates has been described as “antirecombinase” activity [Citation81].
Given that Srs2, Mph1 and Sgs1 all restrict crossover formation during mitosis [Citation84,Citation104–Citation106]; it is likely that their activities may be either down regulated or otherwise altered to allow for the formation of crossover recombination products in meiosis. Indeed, it has been shown that the ZMM proteins (including, Zip1, Zip2, Zip3, Zip4–Spo22, Mer3, Msh4 and Msh5, all of which components of the synaptonemal complex) suppress the antirecombinase function of Sgs1 during later stages on recombination [Citation107]. In addition, Rad51 foci are disrupted by overexpression of Srs2 during meiosis, whereas Dmc1 foci remain unaffected [Citation92]. Moreover, recent in vitro studies have shown that Dmc1 filaments block the antirecombinase activity of Srs2 by inhibiting its ATP hydrolysis and ssDNA translocation activities [Citation108]. These studies highlight a new biochemical distinction between Rad51 and Dmc1, namely, Rad51 is highly susceptible to the antirecombinase activity of Srs2 whereas Dmc1 is not [Citation108]. This finding suggests the possibility that that Dmc1 acts as a pro–crossover recombinase by specifically inhibiting the activity of Srs2. Taken together, these studies are beginning to address how the helicases that participate in HR are differentially regulated to ensure physiologically appropriate recombination outcomes during mitosis and meiosis.
Perspectives
There are many differences between meiotic and mitotic recombination, including differences in the recombinases involved, and well as the participating protein co–factors: such as the Rad51–interacting proteins Rad52, Rad54, Hed1, Srs2; and the Dmc1–interacting proteins Mie5–Sae3, Hop2–Mnd1 and Rdh54 [Citation33,Citation109]. In addition to differences in protein co–factors, there is also emerging evidence for differences in Rad51 and Dmc1 biochemical properties, such as the Dmc1 requirement for Ca [Citation2,Citation36,Citation43] and its ability to stabilize mismatched heteroduplex intermediates [Citation45], however, the biological implications of these differences are still unknown. Less obvious are the reasons why these two distinct recombinases might exist in the first place, as Rad51 and Dmc1 appear to be catalyzing fundamentally the same DNA strand exchange reaction [Citation18,Citation110]. Similarly, it is not at all clear what early evolutionary pressures may have led to emergence of the separate Rad51 and Dmc1 recombinase lineages within the Rad51/RecA family of recombinases [Citation32]. Interestingly, there are eukaryotes that have lost Dmc1 and its associated co–factors, including ecdysozoans such as Drosophilia sp. and Caenorhabditis sp., as well as fungal species such as Ustilago maydis, Sodaria macrospora and Neurospora crassa [Citation33]. Thus, comparison of the meiotic programs and biochemical properties of Rad51 from these organisms with those of organisms that follow to more typical two–recombinase paradigm may provide insights into these evolutionary questions. One interesting experiment would be to simply replace the mitotic HR machinery (Rad51 plus its associated factors) with the meiotic HR machinery (Dmc1 and its associated factors), and vice versa, and ask how mitotic and meiotic recombination are affected. Although this sounds simple, any outcome would have to be interpreted with the understanding that the intra-nuclear chromosome organization is completely different during mitotic versus meiotic recombination, and these differences are likely to have a profound impact on reaction outcomes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. 2015;7:a016618.
- Haber JE. Mating-type genes and MATSwitching in saccharomyces cerevisiae. Genetics. 2012;191:33–64.
- Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet. 2014;48:187–214.
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384.
- Diaz RL, Alcid AD, Berger JM, et al. Identification of residues in yeast spo11p critical for meiotic DNA double-strand break formation. Mol Cell Biol. 2002;22:1106–1115.
- Mimitou EP, Yamada S, Keeney S. A global view of meiotic double-strand break end resection. Science. 2017;355:40–45.
- Symington LS. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb Perspect Biol. 2014;6:195–212.
- Cejka P, Cannavo E, Polaczek P, et al. DNA end resection by DNA2–sgs1–RPA and its stimulation by Top3–rmi1 and Mre11–rad50–xrs2. Nature. 2010;467:112–116.
- Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell. 2013;50:589–600.
- Lisby M, Barlow JH, Burgess RC, et al. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713.
- Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092.
- Brown MS, Grubb J, Zhang A, et al. Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLoS Genet. 2015;11:e1005653.
- Gasior SL, Wong AK, Kora Y, et al. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221.
- Ferrari SR, Grubb J, Bishop DK. The Mei5-Sae3 protein complex mediates dmc1 activity in saccharomyces cerevisiae. J Biol Chem. 2009;284:11766–11770.
- Tsubouchi H, Roeder GS. The budding yeast Mei5 and Sae3 proteins act together with Dmc1 during meiotic recombination. Genetics. 2004;168:1219–1230.
- Filippo JS, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257.
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750.
- Busygina V, Gaines WA, Xu Y, et al. Functional attributes of the s. cerevisiae meiotic recombinase Dmc1. DNA Repair (Amst). 2013;12:707–712.
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70.
- Hunter N, Kleckner N. The single-end invasion. Cell. 2001;106:59–70.
- Morrical SW. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol. 2015;7:a016444.
- Kowalczykowski SC. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2015;711.
- Bzymek M, Thayer NH, Oh SD, et al. Double holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941.
- Martini E, Diaz RL, Hunter N, et al. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295.
- Shinohara M, Sakai K, Shinohara A, et al. Crossover interference in saccharomyces cerevisiaeRequires a TID1/RDH54and DMC1 dependent pathway. Genetics. 2003;163:1273–1286.
- Wang S, Zickler D, Kleckner N, et al. Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle. 2015;14:305–314.
- Youds JL, Boulton SJ. The choice in meiosis – defining the factors that influence crossover or non-crossover formation. J Cell Sci. 2011;124:501–513.
- Moens PB, Kolas NK, Tarsounas M, et al. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J Cell Sci. 2002;115:1611–1622.
- Lao JP, Hunter N. Trying to avoid your sister. PLoS Biol. 2010;8:e1000519.
- Sheridan S, Bishop DK. Red-hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 2006;20:1685–1691.
- Bishop DK, Park D, Xu L, et al. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456.
- Lin Z, Kong H, Nei M, et al. Origins and evolution of the recA/RAD51gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Nat Acad Sci. 2006;103:10328–10333.
- Brown MS, Bishop DK. DNA strand exchange and reca homologs in meiosis. Cold Spring Harb Perspect Biol. 2015;7:a016659.
- Sheridan SD, Yu X, Roth R, et al. A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 2008;36:4057–4066.
- Cloud V, Chan Y-L, Grubb J, et al. Dmc1 catalyzes interhomolog joint molecule formation in meiosis with Rad51 and Mei5-Sae3 as accessory factors. Science. 2012;337:1222–1225.
- Chan Y-L, Brown MS, Qin D, et al. The third exon of the budding yeast meiotic recombination gene HOP2 is required for calcium-dependent and recombinase Dmc1-specific stimulation of homologous strand assimilation. J Biol Chem. 2014;289:18076–18086.
- Henry JM, Camahort R, Rice DA, et al. Mnd1/Hop2 facilitates Dmc1-dependent interhomolog crossover formation in meiosis of budding yeast. Mol Cell Biol. 2006;26:2913–2923.
- Hong-Rae C, Yoon-Ju K, Soo-Gil H, et al. Hop2 and Sae3 are required for dmc1-mediated double-strand break repair via homolog bias during meiosis. Mol Cells. 2016;39:550–556.
- Leu J-Y, Chua PR, Roeder GS. The meiosis-specific hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386.
- Chi P, Kwon Y, Moses DN, et al. Functional interactions of meiotic recombination factors Rdh54 and Dmc1. DNA Repair (Amst). 2009;8:279–284.
- Nimonkar AV, Dombrowski CC, Siino JS, et al. Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J Biol Chem. 2012;287:28727–28737.
- Lee M-H, Chang Y-C, Hong EL, et al. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J Biol Chem. 2005;280:40980–40984.
- Crickard JB, Kaniecki K, Kwon Y, et al. Spontaneous self-segregation of Rad51 and Dmc1 DNA recombinases within mixed recombinase filaments. J Biol Chem. 2018;293:4191–4200.
- Qi Z, Redding S, Lee JY, et al. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015;160:856–869.
- Lee JY, Terakawa T, Qi Z, et al. Base triplet stepping by the Rad51/RecA family of recombinases. Science. 2015;349:977–981.
- Lee JY, Steinfeld JB, Qi Z, et al. Sequence imperfections and base triplet recognition by the Rad51/RecA family of recombinases. J Biol Chem. 2017;292:11125–11135.
- Greene EC. DNA sequence alignment during homologous recombination. J Biol Chem. 2016;291:11572–11580.
- Qi Z, Greene EC. Visualizing recombination intermediates with single-stranded DNA curtains. Methods. 2016;105:62–74.
- Hong S, Sung Y, Yu M, et al. The logic and mechanism of homologous recombination partner choice. Mol Cell. 2013;51:440–453.
- Busygina V, Saro D, Williams G, et al. Novel attributes of hed1 affect dynamics and activity of the Rad51 presynaptic filament during meiotic recombination. J Biol Chem. 2012;287:1566–1575.
- Tsubouchi H, Roeder GS. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775.
- Daley JM, Niu H, Sung P. Roles of DNA helicases in the mediation and regulation of homologous recombination. Adv Exp Med Biol. 2013;767:185–202.
- Mazin AV, Mazina OM, Bugreev DV, et al. Rad54, the motor of homologous recombination. DNA Repair (Amst). 2010;9:286–302.
- Solinger JA, Kiianitsa K, Heyer W-D. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51: dsDNAFilaments. Mol Cell. 2002;10:1175–1188.
- Solinger JA, Lutz G, Sugiyama T, et al. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament1. J Mol Biol. 2001;307:1207–1221.
- Raoul Tan TL, Kanaar R, Wyman C. Rad54, a Jack of all trades in homologous recombination. DNA Repair (Amst). 2003;2:787–794.
- Renkawitz J, Lademann CA, Jentsch S. Mechanisms and principles of homology search during recombination. Nat Rev Mol Cell Biol. 2014;15:369–383.
- Van Komen S, Petukhova G, Sigurdsson S, et al. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell. 2000;6:563–572.
- Wolner B, Peterson CL. ATP-dependent and ATP-independent Roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J Biol Chem. 2005;280:10855–10860.
- Heyer W-D, Li X, Rolfsmeier M, et al. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125.
- Li X, Zhang X-P, Solinger JA, et al. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35:4124–4140.
- Wright WD, Heyer W-D. Rad54 functions as a Heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D-loop formation in homologous recombination. Mol Cell. 2014;53:420–432.
- Liu Y, Gaines WA, Callender T, et al. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand Breaks. PLoS Genet. 2014;10:e1004005.
- Niu H, Wan L, Busygina V, et al. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell. 2009;36:393–404.
- Callender TL, Laureau R, Wan L, et al. Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet. 2016;12:e1006226.
- Hollingsworth NM. Phosphorylation and the creation of interhomolog bias during meiosis in yeast. Cell Cycle. 2010;9:436–437.
- Niu H, Li X, Job E, et al. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol. 2007;27:5456–5467.
- Busygina V, Sehorn MG, Shi IY, et al. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 2008;22:786–795.
- Crickard JB, Kaniecki K, Kwon Y, et al. Regulation of Hed1 and Rad54 binding during maturation of the meiosis‐specific presynaptic complex. Embo J. 2018;37:e98728.
- Hollingsworth NM. Mek1/Mre4 is a master regulator of meiotic recombination in budding yeast. Microb Cell. 2016;3:129–131.
- Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70.
- Neiman AM. Sporulation in the Budding yeast saccharomyces cerevisiae. Genetics. 2011;189:737–765.
- Say AF, Ledford LL, Sharma D, et al. The budding yeast Mei5-Sae3 complex interacts with Rad51 and preferentially binds a DNA fork structure. DNA Repair (Amst). 2011;10:586–594.
- Hayase A, Takagi M, Miyazaki T, et al. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940.
- Pezza RJ, Camerini-Otero RD, Bianco PR. Hop2-Mnd1 Condenses DNA to stimulate the synapsis phase of DNA strand exchange. Biophys J. 2010;99:3763–3772.
- Pezza RJ, Voloshin ON, Vanevski F, et al. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–1766.
- Kang H-A, Shin H-C, Kalantzi A-S, et al. Crystal structure of Hop2–mnd1 and mechanistic insights into its role in meiotic recombination. Nucleic Acids Res. 2015;43:3841–3856.
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57.
- Kohl KP, Sekelsky J. Meiotic and mitotic recombination in meiosis. Genetics. 2013;194:327–334.
- Symington LS, Rothstein R, Lisby M. Mechanisms and regulation of mitotic recombination in saccharomyces cerevisiae. Genetics. 2014;198:795–835.
- Ira G, Malkova A, Liberi G, et al. Srs2 and Sgs1–top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411.
- Liu J, Ede C, Wright WD, et al. Srs2 promotes synthesis-dependent strand annealing by disrupting DNA polymerase δ-extending D-loops. eLife. 2017;6:e22195.
- M-C M-K, Khan MM, Schott J, et al. Mechanistic view and genetic control of DNA recombination during meiosis. Mol Cell. 2018;70:9–20.e6.
- Jain S, Sugawara N, Mehta A, et al. Sgs1 and Mph1 helicases enforce the recombination execution checkpoint during DNA double-strand break repair in saccharomyces cerevisiae. Genetics. 2016;203:667–675.
- Stafa A, Donnianni RA, Timashev LA, et al. Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics. 2014;196:1017–1028.
- Mazón G, Symington LS. Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination intermediates. Mol Cell. 2013;52:63–74.
- Rong L, Palladino F, Aguilera A, et al. The hyper-gene conversion Hpr5-1 mutation of saccharomyces cerevisiae is an allele of the Srs2/Radh Gene. Genetics. 1991;127:75–85.
- Niu H, Klein HL. Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair. FEMS Yeast Res. 2017;17:fow111.
- Antony E, Tomko EJ, Xiao Q, et al. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115.
- Burgess RC, Lisby M, Altmannova V, et al. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J Cell Biol. 2009;185:969–981.
- Krejci L, Van Komen S, Li Y, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305.
- Sasanuma H, Furihata Y, Shinohara M, et al. Remodeling of the Rad51 DNA strand-exchange protein by the Srs2 Helicase. Genetics. 2013;194:859–872.
- Veaute X, Jeusset J, Soustelle C, et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309.
- Xue X, Sung P, Zhao X. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev. 2015;29:1777–1788.
- Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair (Amst). 2010;9:224–236.
- Prakash R, Satory D, Dray E, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79.
- Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA Repair. Annu Rev Genet. 2010;44:393–417.
- Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol. 2016;51:195–212.
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774.
- Niu H, Chung W-H, Zhu Z, et al. Mechanism of the ATP-dependent DNA end resection machinery from S. cerevisiae. Nature. 2010;467:108–111.
- Ashton TM, Mankouri HW, Heidenblut A, et al. Pathways for holliday junction processing during homologous recombination in saccharomyces cerevisiae. Mol Cell Biol. 2011;31:1921–1933.
- Rockmill B, Fung JC, Branda SS, et al. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr Biol. 1954–62;13: 22.
- Fasching CL, Cejka P, Kowalczykowski SC, et al. Top3-Rmi1 dissolve Rad51-mediated D-loops by a topoisomerase-based mechanism. Mol Cell. 2015;57:595–606.
- Mitchel K, Lehner K, Jinks-Robertson S. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet. 2013;9:e1003340.
- Prakash R, Satory D, Dray E, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79.
- Štafa A, Donnianni RA, Timashev LA, et al. Template switching during break-induced replication is promoted by the mph1 helicase in saccharomyces cerevisiae. Genetics. 2014;196:1017–1028.
- Jessop L, Rockmill B, Roeder GS, et al. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2006;2:e155.
- Crickard JB, Kaniecki K, Kwon Y, et al. Meiosis-specific recombinase Dmc1 is a potent inhibitor of the Srs2 antirecombinase. Proc Nat Acad Sci. 2018;115: E10041–E10048. [Epub ahead of print].
- Crickard JB, Greene EC. Biochemical attributes of mitotic and meiotic presynaptic complexes. DNA Repair (Amst). 2018;71: 148–157. [Epub ahead of print].
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243.
