ABSTRACT
Emerging research has suggested that miRNAs play a significant role in oncogenesis and tumor progression by regulating multiple molecular pathways. Here, we investigated miR-300, which inhibited bladder cancer (BCa) migration by regulating the SP1/MMP9 pathway. miR-300, belonging to the DLK1-DIO3 miRNA cluster, is frequently expressed at lower levels in BCa tissue than in adjacent normal tissue due to DNA methylation. Reinforced expression of miR-300 significantly suppressed the migration of BCa cells. We carried out a search of online databases to predict potential targets of miR-300. Further studies determined that miR-300 directly targeted SP1 and suppressed its expression by specifically binding to its 3ʹ-untranslated region. Meanwhile, downregulated MMP9 may be the final effector of BCa cell mobility. Small interference RNAs silencing SP1 phenocopied the effects of miR-300 overexpression, while restoration of SP1 expression partially rescued the inhibition of metastasis induced by miR-300 overexpression in BCa cells. In conclusion, we unveiled a miR-300/SP1/MMP9 pathway in BCa. These findings demonstrate that miR-300 is a promising tumor suppressor in BCa.
KEYWORDS:
Introduction
Bladder cancer (BCa) has become one of the most common cancers worldwide and ranks as the ninth most frequently diagnosed malignancy. In 2012, approximately 430,000 new BCa cases and 165,000 deaths were reported globally. Additionally, the incidence rate was approximately 50% higher in more developed regions than in less developed regions [Citation1]. According to the most advanced data in the USA, BCa is the 4th most common cancer in men and the 11th most common in women [Citation2]. The incidence of BCa is approximately three to four times higher in men than in women [Citation3]. A series of factors including smoking, diabetes and drug use could contribute to the incidence of BCa [Citation4,Citation5]. However, even though BCa is common, it is not well treated, and recurrence is high in both non-muscle-invasive and muscle-invasive BCa after surgical management [Citation6,Citation7]. Therefore, we urgently need a novel and efficient therapeutic strategy to meaningfully improve the outcome of BCa patients.
MicroRNAs (miRNAs) are endogenous, short (19–22 nucleotides) non-coding RNAs that play a role in gene regulation by sequence-specifically binding the 3ʹ-untranslated region (3ʹ-UTR) of a target mRNA [Citation8]. Emerging discoveries of disordered miRNAs have been reported to accelerate the progression and metastasis of cancer, including BCa. Our team has previously identified a series of miRNAs, including miR-26a, miR-101, miR-124-3p, miR-320c, miR-409-3p, miR-490-5p, miR-576-3p, miR-433, miR-148a and miR-323a-3p, involved in the progression of BCa [Citation9–Citation18].
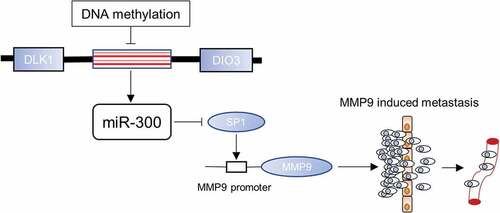
To further investigate the role of miRNAs in the pathogenesis of BCa, we reviewed some published studies on miRNA in human BCa, and the DLK1-DIO3 miRNA cluster captured our attention. The DLK1-DIO3 genomic region is a large miRNA cluster that contains 53 miRNAs in the forward strand and one in the reverse strand [Citation19]. Many of these miRNAs are abnormally expressed and may be involved in the pathogenesis of BCa. Our previous work revealed that miR-323a-3p, miR-433 and miR-409 transcribed from this cluster acted as tumor suppressors [Citation16–Citation18]. miR-300, belonging to the DLK1-DIO3 miRNA cluster, was previously reported to be associated with human osteosarcoma, pancreatic cancer, non-small cell lung cancer, glioblastoma, and oral squamous cell carcinoma [Citation20–Citation24]. However, to the best of our knowledge, the role miR-300 in BCa and its mechanism remains unresolved. In this study, we determined that miR-300 served as a tumor suppressor and inhibited the metastasis of BCa via the SP1/MMP9 pathway. SP1 was confirmed to be the direct target of miR-300 and consequently regulated the expression of MMP9. Thus, a novel mechanism of the miR-300/SP1/MMP9 pathway was established in BCa.
Results
miR-300 is downregulated in BCa
Quantitative real-time PCR (qRT-PCR) was performed to detect the expression levels of miR-300 in BCa cell lines, and the results showed a lower expression level of miR-300 in two BCa cell lines (T24 and UM-UC3) compared to that in the normal bladder cell line (SV-HUC-1) (). We further detected the expression levels of miR-300 in ten pairs of human BCa tissues and adjacent normal tissues by qRT-PCR (). Clinical data from ten patients are shown in Supplementary Table 1. In general, miR-300 expression was lower in the cancerous tissues than in the adjacent normal tissues (8 of 10 pairs of tissue showed a downregulated pattern). In addition, we detected miR-300 expression in BCa tissue microarray (TMA) with CISH method and the result demonstrated that miR-300 was significantly downregulated in BCa tissues than adjacent para-tumor tissues (,). Kaplan-Meier survival analysis revealed that upregulation of miR-300 was significantly associated with high overall survival rate of BCa (). Considering these results, we hypothesized that miR-300 might be a substantial tumor suppressor in BCa.
Figure 1. miR-300 is generally downregulated in bladder cancer tissue. (a) The relative expression levels of miR-300 in T24, UM-UC3 and SV-HUC-1 cells was confirmed by qRT-PCR. (b) The relative expression levels of miR-300 in ten pairs of bladder cancer patient tissue were confirmed by qRT-PCR. (c) The relative expression levels of miR-300 were significantly upregulated after treatment of demethylation agent 5-aza-CdR in UM-UC3 cells. Error bars represent the S.D. obtained from three independent experiments; *P < 0.05. (d) miR-300 was significantly downregulated in BCa tissues than adjacent para-tumor tissues in TMA with CISH method; *P < 0.05. (e) Representative graph of BCa tissues and adjacent para-tumor tissues in TMA with CISH method. (f) Kaplan-Meier survival analysis revealed that upregulation of miR-300 was significantly associated with high overall survival rate of BCa.
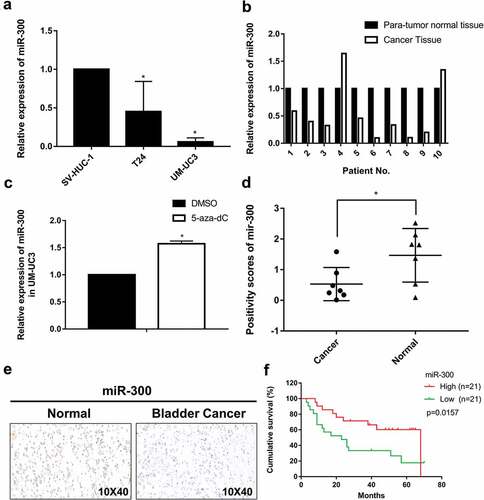
Next, we treated UM-UC3 cells with 5-aza-2′-deoxycytidine (5-aza-CdR), a DNA methyltransferase inhibitor, to identify whether upstream DNA methylation participates in the downregulation of miR-300. As expected, the expression of miR-300 was significantly increased after 5-aza-CdR treatment ()). In our previous study on miR-323-3p and miR-433, which are also located in the imprinted DLK1-DIO3 domain, we confirmed upstream DNA methylation by bisulfite-sequencing PCR [Citation16,Citation17]. These results support our hypothesis that DNA methylation may play a key role in the downregulation of miR-300.
Overexpression of miR-300 inhibits the migration of BCa by downregulating MMP9
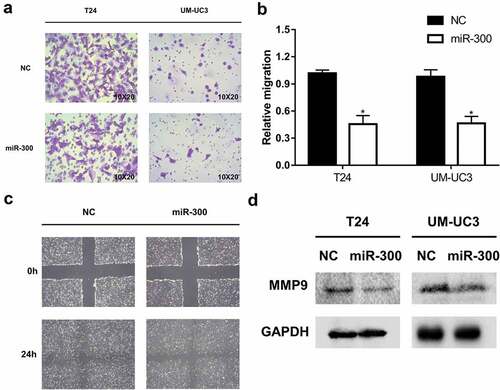
Next, we further explored whether and how miR-300 acted as a tumor suppressor in BCa. After transfection, we conducted the CCK-8 and colony formation assays to explore the effects of miR-300 on cell proliferation, but no significant results were found. However, we found a significant inhibition of cell mobility resulting from miR-300 overexpression, and this result was confirmed by transwell and wound healing assays (-). Our previous study on miR-433 and miR-323-3a, located in the imprinted DLK1-DIO3 domain, like miR-300, revealed an inhibition of metastasis via EMT progression [Citation16,Citation17]. Therefore, we first detected whether EMT progression was also involved in the miR-300-induced inhibition of metastasis. However, no significant changes in EMT protein regulation were found after the overexpression of miR-300 in BCa cells. We next reviewed our previous studies on the imprinted DLK1-DIO3 domain carefully and found that the miR-409-3p, located in the imprinted DLK1-DIO3 domain, inhibited BCa metastasis via downregulating MMP9. Interestingly, we also discovered a significant decrease in the level of MMP9 protein expression after upregulation of miR-300 (). MMP9 is broadly reported to be closely associated with cancer metastasis [Citation25–Citation27]. The downregulation of MMP9 explained the detected suppression of BCa cell mobility after overexpression of miR-300.
Figure 2. miR-300 significantly inhibits the migration of bladder cancer cells. T24 and UM-UC3 cells were transfected by miR-300 mimics or NC (50 nM). (a) The representative micrographs of trans-well assays in T24 and UM-UC3 cells after transfection. Photographs of trans-well assays were obtained under 20× objective. (b) The representative micrographs of trans-well assays were calculated. Error bars represent the S.D. obtained from three independent experiments; *P < 0.05. (c) The wound healing assay revealed a significant inhibition of T24 cell mobility. (d) The Western blot assay confirmed downregulation of MMP9 at protein level in T24 and UM-UC3 cell lines.
miR-300 downregulates SP1 expression by targeting its 3′UTR
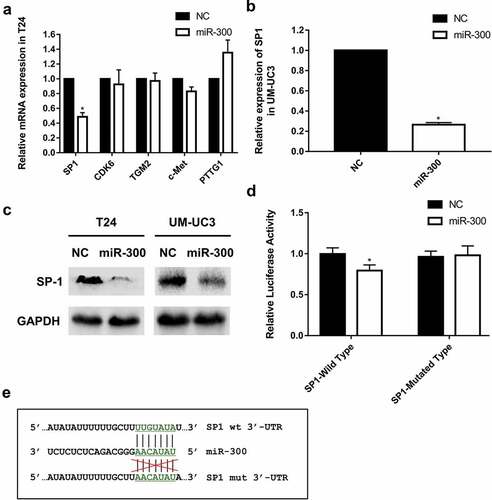
miRNAs can specifically bind to the 3ʹ-UTR of target mRNAs and reduce the expression of target genes. Therefore, we searched online databases (TargetScan, http://www.targetscan.org and MirDatabase, http://www.mirdb.org) to predict the potential targets of miR-300. Five candidate target genes were identified by bioinformatics prediction and their expression levels in T24 cells transfected with miR-300 were detected by qRT-PCR (). The results revealed only one candidate target, SP1, that was downregulated at the mRNA level, and we subsequently confirmed that SP1 was also downregulated in miR-300-overexpressed UM-UC3 cells by qRT-PCR ().
Figure 3. SP1 is a direct target of miR-300. (a-b) A significant downregulation of SP1 was detected by qRT-PCR assay in T24 and UM-UC3 cell lines after overexpression of miR-300. (c) The Western blot assay revealed that miR-300 significantly repressed protein expression of SP1. (d) A Dual-luciferase reporter assay demonstrated that miR-300 significantly inhibited the luciferase activity of vectors carrying 3ʹ-UTR of SP1 in T24 cells. Error bars represent the S.D. obtained from three independent experiments; *P < 0.05. (e) The directly binding sites of miR-300 in the 3ʹUTR of SP1 were mutated as presented.
We next conducted a luciferase reporter assay to confirm the direct interaction between miR-300 and the candidate target SP1. The 3ʹ-UTR of SP1 was cloned into pmirGLO Dual-Luciferase miRNA Target Expression Vectors. We detected that the relative luciferase activity of SP1 was significantly decreased when miR-300 was overexpressed in T24 cells and, as expected, the luciferase activity of the mutated miR-300 targeting site did not decline upon transfection with miR-300 ()). In accordance with the luciferase reporter assays, we detected a significantly reduced protein expression of SP1 in miR-300-transfected cells by Western blot analysis (). These results indicated that miR-300 might suppress MMP9-induced BCa migration by directly binding to the 3ʹ-UTR of SP1. The targeted and mutated sequences of miR-300 are shown as an image ().
Knockdown of SP1 inhibits migration in BCa and downregulates MMP9
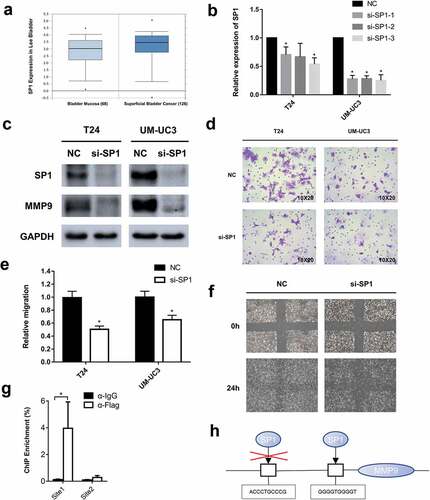
SP1 is a key transcription factor that participates in tumor progression, especially migration. Though SP1 mutations and overexpression are observed in many malignancies, the exact role of SP1 in BCa remains unclear. We first searched the Oncomine database (https://www.oncomine.org/) and found that superficial bladder cancer had a significantly higher SP1 expression level than normal bladder mucosa in a total of 256 samples (, p = 0.018, significant) [Citation28]. This result was consistent with our view that miR-300-targeted SP1 was expressed at higher levels because miR-300 was downregulated in BCa. To verify the precise effect of SP1 in BCa, we used small interference RNAs (siRNAs) to knockdown SP1. The knockdown efficiency of each siRNA was examined by qRT-PCR () and Western-blot assay (). A significant inhibition of migration was detected using the transwell and wound healing assays (-). To further identify the underlying mechanism, a Western blot assay was conducted, and we found that like the SP1 protein, the MMP9 protein was also downregulated after siRNAs were transfected into T24 and UM-UC3 cells (). Previous studies showed that SP1, as a transcription factor, could specifically bind to the promoter site of MMP9 and enhance MMP9 expression [Citation29–Citation31]. So we conducted a ChIP assay in UM-UC3 cells. The prediction of binding region was identified in JARSPAR database (http://jaspar.genereg.net/) [Citation32]. We identified two potential bind-ing regions (site1, GGGGTGGGGT and site2, ACCCTGCCCG) in the promoter zone of MMP9 and the ChIP enrichment was significantly higher in site1 (). A brief figure outlined the mechanism of SP1 specifically binding to the MMP9 promoter (). These results revealed that MMP9 is regulated by SP1 in BCa. Thus, we revealed that knocking down SP1 phenocopied the effect of miR-300 in BCa.
Figure 4. Small interference RNAs were used to knockdown SP1 and a significant inhibition of bladder cancer migration was detected. (a) The data of online database indicates that SP1 is significantly higher expressed in bladder cancer than normal bladder mucosa (p = 0.018, significantly). (b) The knockdown efficiency of each siRNA was determined by qRT-PCR assay. Error bars represent the S.D. obtained from three independent experiments; *P < 0.05. (c) The Western blot assay confirmed downregulation of SP1 and MMP9 at protein levels in T24 and UM-UC3 cells. (d) The representative micrographs of trans-well assay in T24 and UM-UC3 cells after transfected with siRNAs of SP1. Photographs of trans-well assay were obtained under 20× objective. (e) The representative micrographs of trans-well assay were calculated. (f) The wound healing assay revealed a significant inhibition of T24 cell mobility. (g) A ChIP assay revealed that the transcription factor SP1 specifically binds to the promoter zone of MMP9. (h) A sketch outlined the mechanism of SP1 specifically binding to the promoter zone of MMP9.
Overexpressed SP1 partially rescues the miR-300-induced suppression of migration in BCa
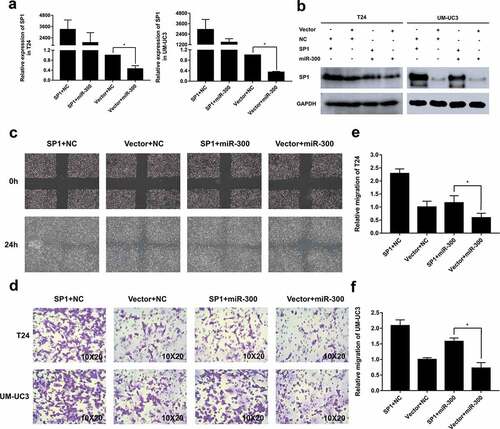
To further confirm the direct association between SP1 and miR-300, we performed a rescue experiment with a SP1-overexpressing plasmid. The transwell and wound healing assays revealed that overexpression of SP1 induced a significant migration of T24 and UM-UC3 cells. More importantly, co-transfection of the SP1 plasmid and miR-300 significantly reversed the inhibited migration induced by miR-300 in the T24 and UM-UC3 cell lines (-). Meanwhile, we examined the relevant SP1 expression levels by qRT-PCR and Western blot analyzes. Exactly as we expected, the changes in SP1 at the mRNA and protein levels were consistent with the phenotype (-). These results indicate that the SP1 downregulation imposed by miR-300 is partially responsible for the miR-300-induced suppression of cell mobility in human BCa and further confirm our conclusion that SP1, as a direct target of miR-300, plays a key role in BCa migration and metastasis.
Figure 5. Overexpression of SP1 partially rescued the miR-300-inhibited mobility of BCa cells. T24 and UM-UC3 cells were co-transfected with pSP1 or vector and miR-300 mimics or NC as displayed. (a) The qRT-PCR assays determined the expression of SP1 at mRNA levels after co-transfection. (b) The Western blot assay confirmed the protein levels of SP1 expression was consistent with qRT-PCR assay. (c) The wound healing assay revealed that the miR-300-induced cell mobility repression was partially rescued by SP1 overexpression. (d) The trans-well assay indicated a consistent trend with the aforesaid assays. (e-f) The relative migration rate of T24 and UM-UC3 cells were calculated. Error bars represent the S.D. obtained from three independent experiments; *P < 0.05.
Discussion
Recently, emerging studies have confirmed the vital role of miRNAs in the development of malignancies. miR-300, as a part of the DLK1-DIO3 genomic imprinted miRNA cluster, is located at chromosomal region 14q32.31 and acts as a tumor suppressor in several different types of human tumors, including lung cancer, pancreatic cancer and laryngeal carcinoma [Citation21,Citation33,Citation34]. However, to the best of our knowledge, its exact role in BCa has not yet been illustrated.
Upstream DNA methylation often plays a key role in silencing downstream gene expression. Many miRNAs, especially those in the imprinted DLK1-DIO3 domain, are reported to be silenced by upstream DNA methylation [Citation35,Citation36]. We previously showed that miR-323-3a in the imprinted DLK1-DIO3 miRNA cluster was downregulated, and the high methylation level of the upstream IG-DMR region was detected using bisulfite-sequencing PCR [Citation17]. Similarly, in our study by Xu. et al, miR-433 in the DLK1-DIO3 domain was also confirmed to be downregulated by upstream DNA methylation [Citation16]. Recently, our team revealed that miR-381-3p, which is neighboring miR-300 in DLK-DIO3 imprinted domain, was also downregulated in BCa because of the hypermethylated status [Citation37]. To clarify why miR-300 is downregulated in bladder cancer, we treated UM-UC3 cells with the demethylation agent 5-aza-CdR, and an obvious upregulation of the miR-300 expression level was detected. These result supports our hypothesis that the downregulation of miR-300 in BCa is induced by upstream DNA methylation.
Next, we performed a gain-of-function experiment. Though miR-300 had no effect on BCa cell proliferation, we clarified that miR-300 functioned as a tumor suppressor by regulating the SP1/MMP9 pathway, as it significantly inhibited the migration of T24 and UM-UC3 cells. Although several studies have depicted the function and mechanism of miR-300, the exact molecular basis of the miR-300-mediated antitumor effect in BCa has not been well elucidated. In this study, we identified and verified that SP1 was a direct target of miR-300. SP1 is a widely characterized transcription factor that often promotes the proliferation, migration and invasion of cancer [Citation38,Citation39]. In our previous study, we characterized miR-124-3p, which indirectly downregulated SP1 and contributed to the metastasis of BCa [Citation15]. These studies support our view that the downregulation of miR-300 abnormally activated SP1 expression and contributed to bladder cancer metastasis.
Matrix metalloproteinases (MMPs), including MMP9, are substantial enzymes that trigger degradation of the basement membrane and extracellular matrix, thereby contributing to cancer migration and metastasis. In this study, the protein level of MMP9 was significantly reduced after the transfection of miR-300 and SP1 siRNAs. It has been proved that SP1, as a stimulatory transcription factor, can specifically bind to the promoter region of MMP9 and plays a critical role in regulating MMP9 expression. In study by Ho et al, the authors used a synthetic promoter containing SP1 transcriptional region and confirm that MMP9 promoter region activity could be regulated by SP1 region [Citation29]. Similarly in study by Lin et al, the authors conducted a luciferase assay to verify that SP1 could bind to the MMP9 promoter [Citation31]. In our study, a ChIP assay was conducted to certify that SP1 specifically binds to the promoter of MMP9 and upregulates the expression of MMP9.These results demonstrated that MMP9 might be the downstream effector protein of miR-300, which appeared to decrease MMP9 expression by targeting SP1 and then inhibiting the ability of cancer metastasis. In another study on miR-892b, the authors also portrayed a SP1/MMP9 pathway in BCa [Citation40]. However, some deficiencies existed in the study: (a) The authors did not clarify the exact function of SP1 in BCa, and (b) the correlation between SP1 and MMP9 was not well elucidated. In our study, we used SP1 siRNAs to determine the function of SP1 in BCa and explicitly noted that SP1 could specifically bind to the promoter region of MMP9 confirmed by the ChIP assay and plays a critical role in regulating MMP9 expression. Nevertheless, we should state that a single miRNA can regulate numerous target genes. It is difficult to identify all the miR-300-mediated signaling pathways and target genes that may have a role in the miR-300-induced inhibition of migration and metastasis in BCa.
In conclusion, our study confirms that miR-300, belonging to the imprinted DLK1-DIO3 domain, is generally downregulated by DNA methylation in bladder cancer. miR-300, which directly targets SP1, exerts an inhibitory effect on BCa cell metastasis via downregulating MMP9. Thus, we unveiled a miR-300/SP1/MMP9 pathway in BCa (). In the future, it may be possible to recruit miR-300 as a novel BCa target for precision therapy on the foundation of the function and mechanism of miR-300 clarified in this study.
Materials and methods
Cell lines and cell culture
T24, UM-UC3 and the normal bladder epithelial cell line SV-HUC-1 were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cell lines were verified by short tandem repeat DNA profiling analysis. T24 cells were cultured in RPMI 1640 medium. UM-UC3 and SV-HUC-1 cells were cultured in MEM. Ten percent heat-inactivated fetal bovine serum (FBS) was added to the above media. All cell lines were cultured at 37°C under a humidified atmosphere of 5% CO2.
Human clinical samples
Paired BCa tissues and adjacent non-tumorous bladder mucosal tissues were obtained from patients undergoing radical cystectomy. The samples were collected between January 2011 and October 2013 at the First Affiliated Hospital of Zhejiang University with informed consent from patients and approval of the Ethics Committee. Tissue samples were immediately frozen in liquid nitrogen until RNA extraction. Patient information is listed in Supplementary Table 1.
Chromogenic in situ hybridization (CISH)
Tissue microarray were purchased from Xinchao Biotech, Shanghai, China, which contained 10 cases with paired tumor and para-tumor tissues and 46 tumor tissues without para-tumor tissue. A miRNA probe was applied as previously described [Citation17]. The probe sequence was listed in Supplementary Table 2.
5-aza-CdR treatment
UM-UC3 cell lines were treated with 5 µM 5-aza-CdR (Sigma, St Louis, MO, USA) for 4 d. qRT-PCR was performed to examine the expression of miR-300.
Reagents and transfection
The RNA duplexes were chemically synthesized by GenePharma Company, Shanghai, China. The miR-300 mimic (named hsa-miR-300, sense; 5′-UAUACAAGGGCAGACUCUCUCU-3′) and the negative control duplex (named NC, sense; 5′-ACTACTGAGTGACAGTAGA-3′), which have no significant homology to any known human sequences, were used for gain-of-function studies. The small interfering RNAs targeting human SP1 mRNA were named as follows:
si-SP1-1 (sense): 5′- CCUGGAGUGAUGCCUAAUAdTdT-3′;
si-SP1-2 (sense):5′- CAGCUUGGUAUCAUCACAAdTdT-3′;
si-SP1-3 (sense): 5′- GGCAGACCUUUACAACUCAdTdT-3′.
The interference efficiency of each siRNA was determined by qRT-PCR. To achieve better knockdown efficiency and fewer off-targets effect, three siRNAs were merged together as a siRNA pool to co-transfect BCa cell lines in the subsequent interference experiments. Oligonucleotide transfection was performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s protocol. FuGENE HD Transfection Reagent (Promega, Madison, WI, USA) was utilized for all plasmid transfections according to the manufacturer’s protocol.
RNA isolation and qRT-PCR
RNA was extracted from cell lines and clinical samples with RNAiso plus (TaKaRa, Kusatsu, Japan) and reverse transcribed into cDNA with the PrimeScript RT reagent Kit and a One-Step PrimeScript miRNA cDNA Synthesis Kit. The expression levels of miRNA and mRNA were detected by qRT-PCR. SYBR Premix Ex Taq (TaKaRa, Kusatsu, Japan) was used to quantify the transcribed cDNA on the ABI 7500 fast real-time PCR System (Applied Biosystems, Carlsbad, USA). Small nuclear RNA U6 and GAPDH mRNA were used as endogenous references to calculate the relative expression of associated genes with the 2−ΔΔCt (delta-delta-Ct algorithm) method. All primers used are listed in Supplementary Table 2.
Western blot analysis
All proteins in this study were extracted from transfected cell lines with RIPA lysis buffer. The relative concentrations were then quantified with a BCA Protein Assay kit. Subsequently, the extracted proteins were loaded onto 10% SDS-polyacrylamide gels and fully electrophoresed. Next, the fully separated proteins were transferred to PVDF membranes with a wet transfer method. After blocking with 5% fat-free milk for 1 h, the membrane was incubated with the primary antibody (at 1:1000 dilution) at 4°C overnight. The membrane was washed with TBS-Tween buffer three times for a total of 30 min and then immersed in secondary antibody (diluted with diluent at a 1:5000 ratio) for 1 h at room temperature. After three washes in TBS-Tween buffer, the protein level was detected with an enhanced chemiluminescence system (Pierce Biotechnology Inc, Rockford, USA). The following primary antibodies were used: rabbit anti-SP1 (Proteintech Group Inc, Rosemont, USA), rabbit anti-MMP9 (Proteintech Group Inc, Rosemont, USA), and rabbit anti-GAPDH (Cell Signaling Technology, Beverly, MA, USA).
Dual-luciferase reporter assay
We designed and ordered the oligonucleotide pairs including the desired miR-300 target region or mutant target region from Sangon, Shanghai, China. After the annealing step, these double-stranded segments were inserted into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) between the SacI and SalI sites. DNA sequencing was employed to verify the insertions. T24 cells were seeded in 24-well plates and co-transfected with 50 nM miR-300 or NC and 100 ng of reporter pmirGLO. The relative luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega) 48 h after transfection.
Chromatin immunoprecipitation assay (ChIP)
A ChIP assay was conducted with SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005 (Cell Signaling Technology, USA) following the protocol of the kit. SP1 (D4C3) Rabbit mAb #9389 (Cell Signaling Technology, USA) was used for the ChIP assay. The primer sequences were listed in Supplementary Table 2
Transwell assay
A transwell assay was used to evaluate cell migration with the use of transwell chambers (Millipore, Boston, MA, USA). After transfection, approximately 5 × 104 T24 cells or 8 × 104 UM-UC3 cells were suspended in 200 µl of serum-free MEM and placed onto the surface layer of the chambers. Then, the entire chamber was placed into a 24-well plate, and 600 µL of medium supplemented with 10% FBS was added to the lower compartment. After incubation for 24 h at 37°C, the chambers were treated with methanol and 0.1% crystal violet. Cells on the upper surface of the membrane were carefully removed using a cotton swab. Photographs were obtained by phase-contrast microscopy (CARL ZEISS, Germany) with a 20× objective.
Wound healing assay
After transfection, cells were grown to 100% confluence in six-well plates, and a micropipette tip was used to make a cross wound. Wound healing was observed after 24 h of culture. Photographs were taken by phase-contrast microscopy (Olympus, Tokyo, Japan)
Statistical analysis
The data are expressed as the mean ± standard deviation (S.D.). Differences between groups were estimated using Student’s t-test. All analyzes were conducted using SPSS 24 software (IBM, Armonk, NY, USA), and significance was defined as a two-tailed p value<0.05.
Supplemental Material
Download MS Word (17.7 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
Additional information
Funding
References
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96–108.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30.
- Dobruch J, Daneshmand S, Fisch M, et al. Gender and bladder cancer: a collaborative review of etiology, biology, and outcomes. Eur Urol. 2016;69:300–310.
- Yan H, Xie H, Ying Y, et al. Pioglitazone use in patients with diabetes and risk of bladder cancer: a systematic review and meta-analysis. Cancer Manag Res. 2018;10:1627–1638.
- Yan H, Ying Y, Xie H, et al. Secondhand smoking increases bladder cancer risk in nonsmoking population: a meta-analysis. Cancer Manag Res. 2018;10:3781–3791.
- Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388:2796–2810.
- McConkey DJ, Choi W, Ochoa A, et al. Intrinsic subtypes and bladder cancer metastasis. Asian J Urol. 2016;3:260–267.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233.
- Hu Z, Lin Y, Chen H, et al. MicroRNA-101 suppresses motility of bladder cancer cells by targeting c-Met. Biochem Biophys Res Commun. 2013;435:82–87.
- Li S, Xu X, Xu X, et al. MicroRNA-490-5p inhibits proliferation of bladder cancer by targeting c-Fos. Biochem Biophys Res Commun. 2013;441:976–981.
- Liang Z, Li S, Xu X, et al. MicroRNA-576-3p inhibits proliferation in bladder cancer cells by targeting cyclin D1. Mol Cells. 2015;38:130–137.
- Lin Y, Chen H, Hu Z, et al. miR-26a inhibits proliferation and motility in bladder cancer by targeting HMGA1. FEBS Lett. 2013;587:2467–2473.
- Wang X, Liang Z, Xu X, et al. miR-148a-3p represses proliferation and EMT by establishing regulatory circuits between ERBB3/AKT2/c-myc and DNMT1 in bladder cancer. Cell Death Dis. 2016;7:e2503.
- Wang X, Wu J, Lin Y, et al. MicroRNA-320c inhibits tumorous behaviors of bladder cancer by targeting Cyclin-dependent kinase 6. J Exp Clin Cancer Res. 2014;33:69.
- Xu X, Li S, Lin Y, et al. MicroRNA-124-3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J Transl Med. 2013;11:276.
- Xu X, Zhu Y, Liang Z, et al. c-Met and CREB1 are involved in miR-433-mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3beta/Snail signaling. Cell Death Dis. 2016;7:e2088.
- Li J, Xu X, Meng S, et al. MET/SMAD3/SNAIL circuit mediated by miR-323a-3p is involved in regulating epithelial-mesenchymal transition progression in bladder cancer. Cell Death Dis. 2017;8:e3010.
- Xu X, Chen H, Lin Y, et al. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells. 2013;36:62–68.
- Benetatos L, Hatzimichael E, Londin E, et al. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70:795–814.
- Chen Z, Zhang W, Jiang K, et al. MicroRNA-300 regulates the ubiquitination of PTEN through the CRL4B(DCAF13) E3 ligase in osteosarcoma cells. Mol Ther Nucleic Acids. 2018;10:254–268.
- Zhang JQ, Chen S, Gu JN, et al. MicroRNA-300 promotes apoptosis and inhibits proliferation, migration, invasion and epithelial-mesenchymal transition via the Wnt/beta-catenin signaling pathway by targeting CUL4B in pancreatic cancer cells. J Cell Biochem. 2018;119:1027–1040.
- Zhang Y, Guo Y, Yang C, et al. MicroRNA-300 targets hypoxia inducible factor-3 alpha to inhibit tumorigenesis of human non-small cell lung cancer. Neoplasma. 2017;64:554–562.
- Zhou F, Li Y, Hao Z, et al. MicroRNA-300 inhibited glioblastoma progression through ROCK1. Oncotarget. 2016;7:36529–36538.
- Lin CC, Chen PC, Lein MY, et al. WISP-1 promotes VEGF-C-dependent lymphangiogenesis by inhibiting miR-300 in human oral squamous cell carcinoma cells. Oncotarget. 2016;7:9993–10005.
- Jin H, Xie Q, Guo X, et al. p63alpha protein up-regulates heat shock protein 70 expression via E2F1 transcription factor 1, promoting Wasf3/Wave3/MMP9 signaling and bladder cancer invasion. J Biol Chem. 2017;292:15952–15963.
- Luo KW, Wei C, Lung WY, et al. EGCG inhibited bladder cancer SW780 cell proliferation and migration both in vitro and in vivo via down-regulation of NF-kappaB and MMP-9. J Nutr Biochem. 2017;41:56–64.
- Gao Y, Guan Z, Chen J, et al. CXCL5/CXCR2 axis promotes bladder cancer cell migration and invasion by activating PI3K/AKT-induced upregulation of MMP2/MMP9. Int J Oncol. 2015;47:690–700.
- Lee JS, Leem SH, Lee SY, et al. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J Clin Oncol. 2010;28:2660–2667.
- Ho HY, Lin CW, Chien MH, et al. Melatonin suppresses TPA-induced metastasis by downregulating matrix metalloproteinase-9 expression through JNK/SP-1 signaling in nasopharyngeal carcinoma. J Pineal Res. 2016;61:479–492.
- Wang W, Yang C, Wang XY, et al. MicroRNA-129 and −335 promote diabetic wound healing by inhibiting Sp1-Mediated MMP-9 expression. Diabetes. 2018;67:1627–1638.
- Lin H-Y, Chen Y-S, Wang K, et al. Fisetin inhibits epidermal growth factor–induced migration of ARPE-19 cells by suppression of AKT activation and Sp1-dependent MMP-9 expression. Mol Vis. 2017;23:900–910.
- Khan A, Fornes O, Stigliani A, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D1284.
- He J, Feng X, Hua J, et al. miR-300 regulates cellular radiosensitivity through targeting p53 and apaf1 in human lung cancer cells. Cell Cycle. 2017;16:1943–1953.
- Ge W, Han C, Wang J, et al. MiR-300 suppresses laryngeal squamous cell carcinoma proliferation and metastasis by targeting ROS1. Am J Transl Res. 2016;8:3903–3911.
- Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012;287:42695–42707.
- Lehner B, Kunz P, Saehr H, et al. Epigenetic silencing of genes and microRNAs within the imprinted Dlk1-Dio3 region at human chromosome 14.32 in giant cell tumor of bone. BMC Cancer. 2014;14:495.
- Li J, Ying Y, Xie H, et al. Dual regulatory role of CCNA2 in modulating CDK6 and MET-mediated cell-cycle pathway and EMT progression is blocked by miR-381-3p in bladder cancer. FASEB J. 2018;fj201800667R.
- Daizumoto K, Yoshimaru T, Matsushita Y, et al. A DDX31/Mutant-p53/EGFR axis promotes multistep progression of muscle-invasive bladder cancer. Cancer Res. 2018;78:2233–2247.
- Devanand P, Kim SI, Choi YW, et al. Inhibition of bladder cancer invasion by Sp1-mediated BTG2 expression via inhibition of DNA methyltransferase 1. FEBS J. 2014;281:5581–5601.
- Shin SS, Park SS, Hwang B, et al. MicroRNA-892b influences proliferation, migration and invasion of bladder cancer cells by mediating the p19ARF/cyclin D1/CDK6 and Sp-1/MMP-9 pathways. Oncol Rep. 2016;36:2313–2320.