ABSTRACT
Cervical cancer is a serious threat to women’s health and is the third most common malignancy in women worldwide. Recent studies indicate that the long non-coding RNA CCAT1 plays a role in the malignant behavior of many tumors. However, the role of CCAT1 in cervical cancer is still unknown. Our aim is to evaluate the expression and investigate the regulatory role and potential mechanism of CCAT1 in cervical cancer. CCAT1 expression was measured by qRT-PCR. In addition, CCK-8 assays, colony formation assays, qRT-PCR assays, Transwell assays and xenograft experiments were performed to determine the role of CCAT1 in the proliferation and invasion in cervical cancer cells. The expression of CCAT1 in the cervical cancer tissues was higher than in the adjacent normal tissues. Overexpressing CCAT1 promoted cervical cancer cell proliferation, colony formation, and invasion in vitro. Elevated CCAT1 suppressed miR-181a expression, which was accompanied by an increased expression of MMP14 and HB-EGF. In contrast, knocking down CCAT1 resulted in increased expression of miR-181a, along with decreased expression of MMP14 and HB-EGF. Thus, CCAT1 is a key oncogenic lncRNA associated with cervical cancer and plays a role in promoting cervical cancer cell proliferation and invasion by regulating the miR-181a-5p/MMP14 axis.
KEYWORDS:
Introduction
Cervical cancer is a serious threat to women’s health and is the second most common malignancy in women worldwide [Citation1]. Every year, an estimated 530,000 new cases are diagnosed worldwide, and 85% of the worldwide deaths from cervical cancer occur in underdeveloped or developing countries [Citation2]. Human papillomavirus (HPV) infection results in a majority of the cases of cervical cancer [Citation3], and HPV integration into the human genome plays a role in cervical carcinogenesis [Citation4–Citation7]. HPV integration often occurs in chromosome 8q24, which is near CCAT1 [Citation4]. Thus, the role of CCAT1 in cervical cancer was studied here.
lncRNAs are a class of transcripts longer than 200 nt, and they do not have protein-coding function. Some lncRNAs have been closely correlated to tumorigenesis [Citation8–Citation12]. CCAT1 is one lncRNA located in 8q24. However, the expression and functional role of CCAT1 in cervical cancer are still unknown. Previous study has shown that CCAT1 can induce epithelial‑to‑mesenchymal transition to promote metastasis in lung adenocarcinoma [Citation13]. Also, TP63 and SOX2 were found can cooperatively regulate CCAT1 expression, silencing of CCAT1 substantially reduces cellular growth in different types of squamous cell carcinomas (SCCs) [Citation14]. CCAT1 was also found to be an oncogene in nasopharyngeal carcinoma, gastric cancer and ovarian cancer [Citation15–Citation17]. In our study, we investigate the expression of CCAT1 in cervical cancer and the molecular mechanism of CCAT1 in the progression of cervical cancer.
Results
CCAT1 is upregulated in cervical cancer tissues and cell lines
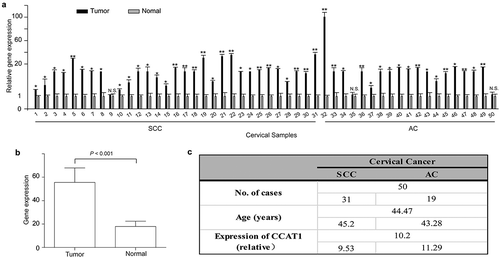
First, we detected the expression of CCAT1 in 50 cervical cancer tissues (including 31 squamous cervical cancer (SCC) and 19 adenocarcinoma (AC) tissues) and its corresponding histologically normal cervical tissues. In 30 of 31 SCCs, the expression of CCAT1 was significantly increased in the cervical cancer tissues compared with the adjacent normal cervical tissues, and in 17 of 19 ACs, similar results were observed. The highest relative expression was 100 fold higher than that in normal tissues ()). Compared with the adjacent normal cervical tissues, the actual expression level of CCAT1 was increased in the cervical cancer tissues (P < 0.0001, )). The statistical analysis is shown in . Also, we detected the expression of CCAT1 in HEK293, SiHa and HeLa cells. The CCAT1 expression in HEK293 was higher than that in SiHa and HeLa (Supplementary FigureS1). These data showed that CCAT1 is upregulated in cervical cancer tissues and cell lines.
Figure 1. CCAT1 is upregulated in cervical cancer tissues. (a) The relative expression of CCAT1 in 50 cervical cancer tissues compared with their adjacent normal cervical tissues. N.S., not significant; *P < 0.05, **P < 0.01. (b) The expression of CCAT1 in the cervical cancer tissues compared with the normal cervical tissues. The expression level of CCAT1 was normalized to GAPDH. (c) Statistical analysis of clinical data. SCC, squamous cell carcinoma; AC, adenocarcinoma.
CCAT1 promotes the proliferation and colony formation of cervical cancer cells
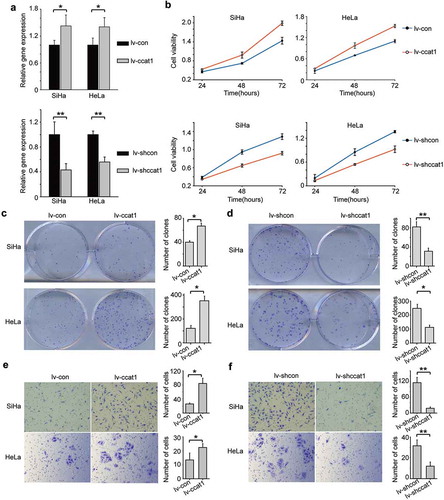
To further investigate the function of CCAT1 in cervical cancer, we transduced SiHa and HeLa cervical cancer cells to over-express the CCAT1 lentivirus (lv-CCAT1) or its control lentivirus (lv-con) or to knock down CCAT1 lentivirus (lv-shCCAT1) or express control shRNA (lv-shcon). Compared with the control cells transduced with lv-con, the relative expression of CCAT1 of the cells transduced with lv-CCAT1 increased ()). Cell Counting Kit-8 (CCK-8) assays showed that the growth of the cells transduced with lv-CCAT1 increased compared to the cells transduced with lv-con ()). In contrast, the cells transduced with lv-shCCAT1 showed decreased CCAT1 expression and slower growth compared with the control cells transduced with lv-shcon (). In addition, we performed a colony formation assay to further study the function of CCAT1 on cervical cancer cell growth. Consistent with the above data, we found that elevated CCAT1 increased the SiHa and HeLa cell colony formation ()), while downregulated CCAT1 decreased the SiHa and HeLa cell colony formation ()). These data indicate that CCAT1 promotes cervical cancer cell proliferation and colony formation.
Figure 2. CCAT1 promotes cervical cancer cell proliferation and invasion. (a) The relative expression of CCAT1 in SiHa and HeLa cells with overexpressed or knocked-down CCAT1 compared the controls. (b) Cell viabilities of the SiHa and HeLa cells at 24, 48 and 72 h after overexpressing or knocking down CCAT1. The cell viabilities were determined by the CCK-8 assay. (c) Clone formation images and statistical analysis of the SiHa and HeLa cells transduced with lv-CCAT1 and lv-con. (d) Clone formation images and statistical analysis of the SiHa and HeLa cells transduced with lv-shCCAT1 and lv-shcon. (e) The invasion ability of the SiHa and HeLa cells transduced with lv-CCAT1 and lv-con or (f) transduced with lv-shCCAT1 and lv-shcon. The error bars represent the mean ± SD of the triplicate experiments. *P < 0.05; **P < 0.01.
CCAT1 promotes invasion by cervical cancer cells
To examine the influence of CCAT1 on cervical cancer cell invasion, we performed Transwell assays using SiHa and HeLa cells. Compared to the controls, elevated CCAT1 promoted SiHa and HeLa cell invasion ()). However, downregulated CCAT1 inhibited SiHa and HeLa cell invasion ()). These data indicate that CCAT1 promotes the invasion of cervical cancer cells.
CCAT1 regulates the expression of miR-181a-5p and matrix metalloproteinase 14 (MMP14)
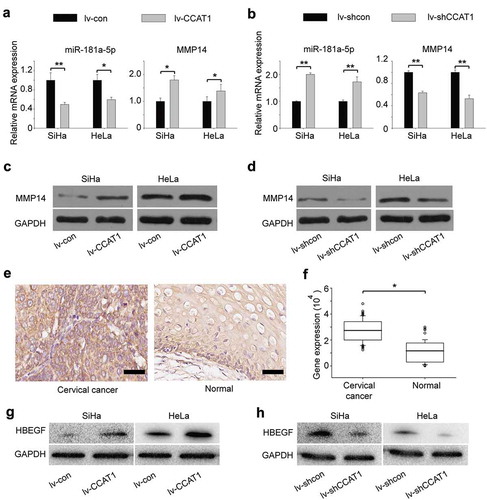
Some lncRNAs contain motifs with complementary sequences to miRNAs [Citation18–Citation21]. To find whether CCAT1 interacts with microRNAs, we queried starBase v.2.0, which predicted that miR-181a-5p interacted with CCAT1 (Supplementary Table 1). MiR-181a-5p negatively regulates the expression of MMP14 [Citation22], which is involved in cell proliferation and invasion [Citation23–Citation25]. To explore the mechanism by which CCAT1 regulated cell proliferation and invasion, we measured the expression of miR-181a-5p using qRT-PCR. We found that when CCAT1 was upregulated, the expression of miR-181a-5p decreased, along with an upregulation of MMP14 ()). In contrast, when CCAT1 was downregulated, the expression of miR-181a-5p increased, along with a downregulation of MMP14 ()). The protein expression of MMP14 was consistent with the RNA level ( and Supplementary Table 2). We also measured the activity of MMP14. We found when CCAT1 was upregulated, the MMP14 activity in SiHa was increased; while when CCAT1 was downregulated, the MMP14 activity in SiHa was decreased (Supplementary FigureS2A). Similar results were detected in HeLa cells (Supplementary FigureS2B). Furthermore, we examined the expression of MMP14 in 60 cervical cancer tissues and 60 adjacent normal tissues. The expression of MMP14 in cervical cancer tissues was much higher than in normal cervical tissues (), indicating that high expression of MMP14 is associated with cervical cancer, which was consistent with other researchers [Citation22,Citation26]. These data indicate that the CCAT1 expression level is negatively correlated with miR-181a-5p and further regulates the expression of MMP14. Additionally, we found that the expression of HB-EGF increased when CCAT1 was upregulated, while the expression of HB-EGF decreased when CCAT1 was downregulated, which was consistent with other researchers’ findings that MMP14 could regulate the expression of HB-EGF () [Citation23,Citation27].
Figure 3. CCAT1-regulated expression of miR-181a-5p and MMP14 in SiHa and HeLa cells. (a) The relative gene expression of miR-181a-5p and MMP14 in the SiHa and HeLa cells transduced with the lv-CCAT1 or (b) lv-shCCAT1 lentivirus and their respective control lentiviruses. (c) Protein expression of MMP14 in the cells transduced with lv-CCAT1 and lv-con. (d) Protein expression of MMP14 in the cells transduced with lv-shCCAT1 and lv-shcon. (e) Protein expression and (f) statistical analysis of MMP14 in cervical cancer tissues and normal cervical tissues. (g) Protein expression of HB-EGF in the cells transduced with lv-CCAT1 and lv-con. (h) Protein expression of HB-EGF in the cells transduced with lv-shCCAT1 and lv-shcon. Lv-CCAT1 and lv-con, CCAT1 overexpression lentivirus and its control lentivirus, respectively. Lv-shCCAT1 and lv-shcon, CCAT1 knockdown lentivirus and its control lentivirus, respectively.
Up- or downregulated miR-181a-5p reverses the expression of MMP14 in cells infected with lv-CCAT1/lv-shCCAT1
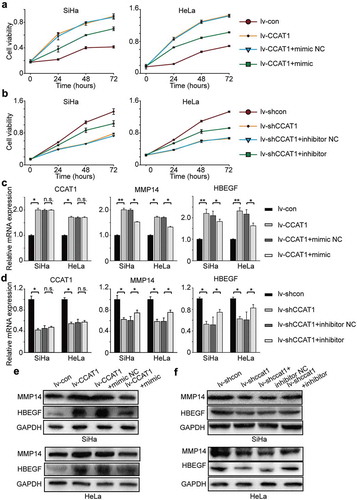
To further confirm that CCAT1 regulates the miR-181a-5p/MMP14 axis, we transduced SiHa and HeLa cells with lv-CCAT1 (lv-CCAT1 cells) and lv-shCCAT1 (lv-shCCAT1 cells) and then transfected with a miR-181a-5p mimic or inhibitor. The lv-CCAT1 cells transfected with the miR-181a-5p mimic showed a decreased growth rate compared with the lv-CCAT1 cells transfected with the miR-181a-5p mimic negative control ()), while the lv-shCCAT1 cells transfected with the miR-181a-5p inhibitor showed an increased growth rate compared with the lv-shCCAT1 cells transfected with the miR-181a-5p inhibitor negative control (). At the mRNA level, the expression of CCAT1 was not significantly different between the lv-CCAT1 cells transfected with the miR-181a-5p mimic negative control and miR-181a-5p mimic, while the expression of MMP14 decreased, as did HB-EGF (). At the protein level, we found that the expression of MMP14 and HB-EGF decreased in the lv-CCAT1 cells transfected with the miR-181a-5p mimic compared with the miR-181a-5p mimic negative control () and Supplementary Table 2). The opposite phenomenon, at both the mRNA and protein levels, was found in the lv-shCCAT1 cells transfected with the miR-181a-5p inhibitor compared with the miR-181a-5p inhibitor negative control () and Supplementary Table 2). These data further indicate that CCAT1 regulated the miR-181a-5p/MMP14 axis to promote cell proliferation and invasion.
Figure 4. The oncogenic activity of CCAT1 partially negatively regulates miR-181a-5p and then modulates MMP14 and HBEGF. (a) Cell viabilities of the SiHa and HeLa cells transduced with lv-CCAT1 and then transfected with miR-181a-5p mimic or (b) lv-shCCAT1 and then transfected with miR-181a-5p inhibitor. The cell viability was measured by the CCK-8 assay. (c) The relative mRNA expression of CCAT1, MMP14 and HB-EGF in SiHa and HeLa cells transduced with lv-CCAT1 and then transfected with miR-181a-5p mimic or (d) transduced with lv-shCCAT1 and then transfected with miR-181a-5p inhibitor. (e) The protein expression of MMP14 and HB-EGF in SiHa and HeLa cells transduced with lv-CCAT1 and then transfected with miR-181a-5p mimic or (f) transduced with lv-shCCAT1 and then transfected with miR-181a-5p inhibitor. Lv-CCAT1 and lv-con, CCAT1 overexpression lentivirus and its control lentivirus, respectively. Lv-shCCAT1 and lv-shcon, CCAT1 knockdown lentivirus and its control lentivirus, respectively.
CCAT1 promotes the tumor growth of cervical cancer cells
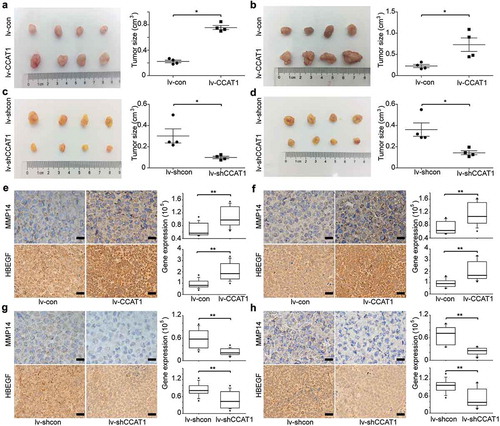
To provide additional evidence that CCAT1 promotes cervical cancer cell proliferation, we used SiHa and HeLa cells transduced with the lentivirus lv-CCAT1, lv-shCCAT1 and their control lentivirus. The cells were then injected subcutaneously into nude mice. We measured the volume of the tumors once a week and euthanized the mice 5 weeks after the cancer cell injection. Compared to the controls, when CCAT1 was upregulated, the tumors of SiHa and HeLa both showed increased growth (). Compared to the control tumors transduced with lv-shcon, the tumors transduced with lv-shCCAT1 both showed slower growth (). Furthermore, the IHC assays demonstrated that the expression levels of MMP14 and HB-EGF increased in the tumors transduced with the lv-CCAT1 lentivirus () and decreased in the tumors transduced with the lv-shCCAT1 lentivirus (). These data confirm that CCAT1 promoted cervical cancer cell tumor growth.
Figure 5. CCAT1 promotes tumor growth of SiHa and HeLa cells. SiHa and HeLa cells were subcutaneously injected in the right flanks of the BALB/c-nu mice. (a) Photographs of the subcutaneously formed tumors and estimated sizes of tumors of SiHa transduced with lv-con/lv-CCAT1 and (c) lv-shcon/lv-shCCAT1. (b) Photographs of the subcutaneously formed tumors and estimated sizes of tumors of HeLa transduced with lv-con/lv-CCAT1 (above) and (d) lv-shcon/lv-shCCAT1. The tumor size of SiHa and HeLa was measured starting 2 weeks after injection. (e) Representative images of IHC staining and a comparison of the protein expression of MMP14 and HB-EGF in xenografts of SiHa and (f) HeLa transduced with lv-con/lv-CCAT1. (g) Representative images of IHC staining and a comparison of the protein expression of MMP14 and HB-EGF in xenografts of SiHa and (h) HeLa transduced with lv-shcon/lv-shCCAT1.
Discussion
Over 70% of the human genome is transcribed into RNA that does not encode proteins, and only a few non-coding RNAs (ncRNAs) have been studied. However, this non-coding part of the genome plays many key roles in the majority of the most important biological processes in cancers. ncRNAs are generally divided into two classes according to their size. Small ncRNAs are 20–200 nucleotides (nt) in size, and long ncRNAs (lncRNAs) are longer than 200 nt and even exceed 100,000 nt [Citation28,Citation29]. These lncRNAs can be categorized as intronic, exonic, or intergenic, according to their proximity to the nearest protein-coding transcripts [Citation30]. They play roles in epigenetic, transcriptional and post-transcription regulation [Citation31,Citation32]. Some lncRNAs are involved in cancerous pathways. For example, HOTAIR is related to breast tumors [Citation33–Citation35] and has been reported to promote cancer metastasis by targeting chromatin repressor Polycomb proteins to specific genomic loci [Citation36] and promoting the proliferation and metastasis of osteosarcoma cells through the AKT/mTOR signaling pathway [Citation37]. CCAT1 is potentially related to laryngeal squamous cancer, osteosarcoma and retinoblastoma [Citation12,Citation38,Citation39]. Additionally, CCAT1 was reported to modulate the sensitivity of paclitaxel in nasopharynx cancers [Citation40]. In our study, CCAT1 was upregulated in cervical cancer compared to normal cervical tissues, which indicates that CCAT1 is related to cervical carcinogenesis.
MicroRNAs (miRNAs) are highly conserved, single-stranded, non-coding RNAs of approximately 21–24 nucleotides that regulate gene expression by the post-transcriptional silencing of specific target RNAs. An increasing number of microRNAs have been shown to be related to cancers. miR-613 impedes the proliferation and invasion of glioma cells [Citation41]; miRNA-454 inhibits non-small-cell lung cancer growth and metastasis by targeting the signal transducer and activator of transcription-3 [Citation42]; and miR-215-5p is a tumor suppressor in colorectal cancer targeting the EGFR ligand epiregulin and its transcriptional inducer HOXB9 [Citation43].
The matrix metalloproteinase (MMP) family proteins are involved in the breakdown of extracellular matrix in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling, as well as in disease processes. MiR-181a-5p is reported to inhibit cell proliferation and invasion by downregulating MMP14 [Citation22,Citation26]. Additionally, MMP14 activity mediates the proteolytic processing and activation of heparin-binding EGF-like growth factor (HB-EGF), stimulating the EGFR signaling pathway to increase proliferation and tumor growth [Citation23], which was related with cervical cancer [Citation44,Citation45].
An increasing number of studies indicate that lncRNAs play carcinogenic roles by regulating some microRNAs. Long non-coding RNAs (lncRNAs) act as competing endogenous RNAs (ceRNAs), where microRNAs (miRNAs) and lncRNAs regulate each other through their binding sites. The long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression [Citation46]. The long non-coding RNA HNF1A-AS1 promotes cell proliferation and invasion by regulating miR-17-5p in non-small-cell lung cancer [Citation11]. The long non-coding RNA MEG3 functions as a ceRNA to regulate ischemic neuronal death by targeting the miR-21/PDCD4 signaling pathway [Citation8].
CCAT1 promotes human retinoblastoma cells and lung cancer through negative regulation of miR-218-5p [Citation12,Citation47]. CCAT1 also promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression [Citation48]. In this study, we demonstrated that lncRNA CCAT1 promoted cervical cancer cell line (SiHa and HeLa) proliferation and invasion by suppressing the expression of miR-181a-5p, resulting in the upregulation of MMP14. Furthermore, we confirmed that CCAT1 promoted cervical cancer tumor growth.
In conclusion, the lncRNA CCAT1 is overexpressed in cervical cancer, and it can promote cervical cancer cell proliferation and invasion by upregulating MMP14 and HB-EGF expression via miR-181a-5p. Our research provides a direction for the study of cervical carcinogenesis and for the targeted treatment of cervical cancer in the clinic.
Materials and methods
Specimens
Fifty fresh cervical cancer tissues and their adjacent normal cervical tissues were obtained from patients diagnosed with cervical cancer in the Department of Obstetrics and Gynecology, Tongji Hospital, Huazhong University of Science and Technology (Supplementary Table 1). The cervical cancer tissue experiments were supervised and approved by the Ethics Committee of Tongji Hospital, Huazhong University of Science and Technology. All the tissue specimens were stored at −80°C until use.
Cell culture
The cervical cancer cell lines SiHa, HeLa and human renal epithelial cell line HEK293 were purchased from ATCC and passaged in our laboratory. The cells were cultured in DMEM supplemented with 10% FBS (Gibco) and 100 U/ml of penicillin and streptomycin (Invitrogen) at 37°C in a humidified incubator with 5% CO2.
RNA extraction from the clinical tissue samples and real-time quantitative reverse transcription–polymerase chain reaction (qRT-PCR)
Total RNA from the cervical cancer tissues and cell lines was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription (RT) was performed with the PrimeScript RT Reagent Kit (Takara, Dalian, China) according to the manufacturer’s protocol. The real-time quantitative polymerase chain reaction (qPCR) was performed with the SYBR PrimeScript RT-PCR Kit (Takara) according to the manufacturer’s instructions. The expression levels of CCAT1, miR-181a-5p and matrix metallopeptidase 14 (MMP14) were calculated with the 2−ΔΔCt method. The normalization controls were U6 (miR-181a-5p) and GAPDH (CCAT1, MMP14 and HB-EGF) mRNA. All the assays were performed in triplicate. The expression level was normalized to the corresponding controls. The sequences of the primers are shown in Table 1. Primers for miR-181a-5p and U6 were purchased from RiboBio Co., Ltd. (Guangzhou, China). The primers were shown as follows: MMP14 forward primers: 5ʹ-GGCTACAGCAATATGGCTACC-3ʹ, reverse primers: 5ʹ-GATGGCCGCTGAGAGTGAC-3ʹ; HB-EGF forward primers: 5ʹ- ATCGTGGGGCTTCTCATGTTT-3ʹ, reverse primers: 5ʹ- TTAGTCATGCCCAACTTCACTTT-3ʹ.
Lentivirus preparation and transduction, miR-181a-5p mimic and inhibitor preparation and transfection
The CCAT1 overexpression lentivirus and knockdown lentivirus were constructed by Shanghai Genechem Co., Ltd. For the CCAT1 overexpression lentivirus (lv-CCAT1), the CCAT1 sequence was cloned into the Ubi-MCS-SV40-EGFP-IRES-puromycin. Its control lentivirus was named lv-con. For the CCAT1 knockdown lentivirus (lv-shCCAT1), the siRNA sequence of CCAT1, AAGCAGGCAGAAAGCCGUAUCUUAA, was cloned into the hU6-MCS-CMV-puror lentivirus. The negative-control scrambled sequence, TTCTCCGAACGTGTCACGT, was cloned into the same lentivirus vector (lv-shcon). The mimic and inhibitor of miR-181a-5p and the negative-control RNAs were purchased from RiboBio Co., Ltd. (Guangzhou, China). All the lentivirus constructs were transduced according to the manufacturer’s instructions, and the miRNA transfections were performed using Lipofectamine 3000 (Thermo Fisher Scientific, Wyman Street, Waltham, MA, USA) according to the manufacturer’s instructions.
Cell proliferation assay
A total of 5 × 10 [Citation3] SiHa or HeLa cells per well were seeded in 96-well plates and were cultured in DMEM. Cell proliferation was determined using the Cell Counting Kit-8 (CCK-8, Dojindo) according to the manufacturer’s manual.
Colony formation assay
A total of 200 SiHa or HeLa cells were plated in triplicate in DMEM with 10% FBS in 6-well plates and were incubated for 2 weeks. The clones were stained with 0.04% crystal violet and photographed. The clones were counted by ImagePro Plus (Version 6.0, Media Cybernetics).
Transwell assay
To explore whether CCAT1 could influence the invasive ability of cervical cancer cells, a Transwell assay was performed using a Transwell chamber with 8.0 μm pores (Corning, USA). The upper chamber was covered by Matrigel (BD, USA), which was diluted with DMEM with no FBS (volume ratio 1:7). Then, cells were cultured in the upper chamber with DMEM without FBS, and the lower chamber contained fresh DMEM with 10% FBS. Forty-eight hours later, cells on the upper chamber were removed and cells from the lower surface were fixed with 4% paraformaldehyde, stained with 0.04% crystal violet. Photographs were taken using cellSens Dimension (version 1.8.1, Olympus). The cells were counted by ImagePro Plus.
Western blot assay
The cells were lysed for 30 min in ice-cold RIPA lysis buffer (Servicebio Inc, Wuhan, China) and a protease inhibitor cocktail (Roche). The primary antibodies used were mouse anti-GAPDH (1:2000, ABclonal), rabbit anti-MMP14 (1:1000, A12743, ABclonal) and rabbit anti-HB-EGF (1:1000, A1695, ABclonal). The proteins were detected with a horseradish peroxidase-conjugated anti-rabbit IG secondary antibody by the ECL system (Bio-Rad). The absorbance values were analyzed by ImagePro Plus.
MMP14 activity assay
MMP14 activity was measured using a SensoLyte 520 MMP14 assay kit (AnaSpec, Fremont, CA) according to the manufacturer’s instructions. Briefly, membranes were resuspended in assay buffer and treated as indicated. Samples were incubated for 2 h at 37 ℃ and then added to MMP14 substrate solution (1:1 volume), and optical fluorescence intensity was measured at 490 nm excitation and 520 nm emission at a 3-h end-point, as indicated.
Xenograft experiments
Four-week-old female BALB/c-nu nude mice were purchased from BEIJING HFK BIOSCIENCE and were housed at the Experimental Animal Center, Tongji Medical College, Huazhong University of Science and Technology (HUST, Wuhan, China). The mice were injected subcutaneously in the right flank with 5 × 10 [Citation6] SiHa and HeLa cells. The mice were randomly divided into groups. The tumor growth was measured using a digital caliper, and the tumor size was calculated using the following formula: L × W [Citation2] × 0.5. The subcutaneous tumors were collected after the mice were euthanized. All the experimental protocols were approved by the Institutional Animal Care and Use Committee of HUST, and our study was carried out in strict accordance with the Guidelines for the Welfare of Animals in Experimental Neoplasia.
Immunohistochemistry staining (IHC)
The mice were euthanized, and the xenografts were isolated and fixed with 4% paraformaldehyde. Paraffin-embedded sections (5 μm) were subjected to IHC staining according to the Proteintech protocol (http://www.ptglab. com/Support/index.aspx). The slides were incubated overnight at 4°C with a rabbit anti-MMP14 (1:100, A12743, ABclonal) and a rabbit anti-HB-EGF primary antibody (1:100, A1695, ABclonal). Antibody detection was performed using DAB. Photographs were taken using cellSens Dimension, and the staining intensity was measured using ImagePro Plus.
Statistical analysis
The data are expressed as the mean ± SD and were analyzed by Student’s t-test. The experiments were performed three times in duplicate. The statistical analyses were performed with GraphPad Prism 5. The statistical analysis for was performed using a one-way ANOVA with P = 0.05 and assuming equal variances.
Supplemental Material
Download MS Word (1.4 MB)Supplemental Material
Download MS Excel (12.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
- PimentaJM, GalindoC, JenkinsD, et al. Estimate of the global burden of cervical adenocarcinoma and potential impact of prophylactic human papillomavirus vaccination. BMC Cancer. 2013;13:553.
- Millikan RC. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst. 1994;86:392–393.
- Hu Z, Zhu D, Wang W, et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47:158–163.
- Jeon S, LambertPF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci U S A. 1995;92:1654–1658.
- Meanwell CA, Cox MF, Blackledge G, et al. HPV 16 DNA in normal and malignant cervical epithelium: implications for the aetiology and behaviour of cervical neoplasia. Lancet. 1987;1:703–707.
- Cancer Genome Atlas Research N, Albert Einstein College of M, Analytical Biological S, et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017; 543: 378–384.
- Yan H, Rao J, Yuan J, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate ischemic neuronal death by targeting miR-21/PDCD4 signaling pathway. Cell Death Dis. 2017;8:3211.
- Yang F, Xue X, Bi J, et al. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445.
- Zhang E, Han L, Yin D, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086–3101.
- Zhang G, An X, Zhao H, et al. Long non-coding RNA HNF1A-AS1 promotes cell proliferation and invasion via regulating miR-17-5p in non-small cell lung cancer. Biomed Pharmacothe. 2017;98:594–599.
- Zhang H, Zhong J, Bian Z, et al. Long non-coding RNA CCAT1 promotes human retinoblastoma SO-RB50 and Y79 cells through negative regulation of miR-218-5p. Biomed Pharmacothe. 2017;87:683–691.
- Lin H, Cheng W, Yan H, et al. Overexpression of the long noncoding RNA CCAT1 promotes metastasis via epithelial-to-mesenchymal transition in lung adenocarcinoma. Oncol Lett. 2018;16:1809–1814.
- Jiang Y, Jiang YY, Xie JJ, et al. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat Commun. 2018;9:3619.
- Dong Y, Yuan H, Jin G. Identification of long non-coding RNA CCAT1 as an oncogene in nasopharyngeal carcinoma. Oncol Lett. 2018;16:2750–2756.
- Eguiluz AG Lattice relaxation at an aluminum surface: self-consistent linear-electronic-response approach. Physical review B, Condens matter 1987; 35:5473–5486.
- Coni P, Madeddu A, Kuqi L, et al. LncRNA colon cancer-associated transcript 1 (CCAT1) in ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:1525–1527.
- Pan X, Li D, Huo J, et al. LINC01016 promotes the malignant phenotype of endometrial cancer cells by regulating the miR-302a-3p/miR-3130-3p/NFYA/SATB1 axis. Cell Death Dis. 2018;9:303.
- Liang L, Xu J, Wang M, et al. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death Dis. 2018;9:372.
- Lin Z, Zhou Z, Guo H, et al. Long noncoding RNA gastric cancer-related lncRNA1 mediates gastric malignancy through miRNA-885-3p and cyclin-dependent kinase 4. Cell Death Dis. 2018;9:607.
- Jiang D, Li H, Xiang H, et al. Long Chain Non-Coding RNA (lncRNA) HOTAIR Knockdown Increases miR-454-3p to Suppress Gastric Cancer Growth by Targeting STAT3/Cyclin D1. Medical science monitor : international medical journal of experimental and . clinical research. 2019;25:1537–1548.
- Li Y, Kuscu C, Banach A, et al. miR-181a-5p inhibits cancer cell migration and angiogenesis via downregulation of matrix metalloproteinase-14. Cancer Res. 2015;75:2674–2685.
- Stawowczyk M, Wellenstein MD, Lee SB, et al. Matrix metalloproteinase 14 promotes lung cancer by cleavage of heparin-binding EGF-like growth factor. Neoplasia. 2017;19:55–64.
- Suetsugu T, Koshizuka K, Seki N, et al. Downregulation of matrix metalloproteinase 14 by the antitumor miRNA, miR-150-5p, inhibits the aggressiveness of lung squamous cell carcinoma cells. Int J Oncol. 2017.
- Nguyen AT, Chia J, Ros M, et al. Organelle specific O-Glycosylation drives MMP14 activation, tumor growth, and metastasis. Cancer Cell. 2017;32: 639–53e6.
- Zhang YH, Wang JJ, Li M, et al. Matrix metallopeptidase 14 plays an important role in regulating tumorigenic gene expression and invasion ability of HeLa cells. Int J Gynecol Cancer. 2016;26:600–606.
- Overland AC, Insel PA. Heterotrimeric G proteins directly regulate MMP14/membrane type-1 matrix metalloprotease: a novel mechanism for GPCR-EGFR transactivation. J Biol Chem. 2015;290:9941–9947.
- Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246.
- Kapranov P, Cawley SE, Drenkow J, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919.
- Lee C, Kikyo N. Strategies to identify long noncoding RNAs involved in gene regulation. Cell Biosci. 2012;2:37.
- Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166.
- Zhu J, Fu H, Wu Y, et al. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci. 2013;56:876–885.
- Ding W, Ren J, Ren H, et al. Long noncoding RNA HOTAIR modulates MiR-206-mediated Bcl-w signaling to facilitate cell proliferation in breast cancer. Sci Rep. 2017;7:17261.
- Hassanzarei S, Hashemi M, Sattarifard H, et al. Genetic polymorphisms of HOTAIR gene are associated with the risk of breast cancer in a sample of southeast Iranian population. Tumour Biol. 2017;39:1010428317727539.
- Li M, Li X, Zhuang Y, et al. Induction of a novel isoform of the lncRNA HOTAIR in Claudin-low breast cancer cells attached to extracellular matrix. Mol Oncol. 2017;11:1698–1710.
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076.
- Li E, Zhao Z, Ma B, et al. Long noncoding RNA HOTAIR promotes the proliferation and metastasis of osteosarcoma cells through the AKT/mTOR signaling pathway. Exp Ther Med. 2017;14:5321–5328.
- <ccat1 promotes laryngeal squamous cell carcinoma cell proliferation and invasion [pmidz27830017].pdf>.
- Zhao J, Cheng L. Long non-coding RNA CCAT1/miR-148a axis promotes osteosarcoma proliferation and migration through regulating PIK3IP1. Acta Biochim Biophys Sin (Shanghai). 2017;49:503–512.
- Wang Q, Zhang W, Hao S. LncRNA CCAT1 modulates the sensitivity of paclitaxel in nasopharynx cancers cells via miR-181a/CPEB2 axis. Cell Cycle. 2017;16:795–801.
- Li Q, Zhou L, Wang M, et al. MicroRNA-613 impedes the proliferation and invasion of glioma cells by targeting cyclin-dependent kinase 14. Biomed Pharmacothe. 2017;98:636–642.
- Liu S, Ge X, Su L, et al. MicroRNA-454 inhibits nonsmall cell lung cancer cells growth and metastasis via targeting signal transducer and activator of transcription-3. Molecular medicine reports 2017.
- Vychytilova-Faltejskova P, Merhautova J, Machackova T, et al. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis. 2017;6:5.
- Murata T, Mizushima H, Chinen I, et al. HB-EGF and PDGF mediate reciprocal interactions of carcinoma cells with cancer-associated fibroblasts to support progression of uterine cervical cancers. Cancer Res. 2011;71:6633–6642.
- Schrevel M, Osse EM, Prins FA, et al. Autocrine expression of the epidermal growth factor receptor ligand heparin-binding EGF-like growth factor in cervical cancer. Int J Oncol. 2017;50:1947–1954.
- Chen DL, Lu YX, Zhang JX, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836–4849.
- Lu L, Xu H, Luo F, et al. Epigenetic silencing of miR-218 by the lncRNA CCAT1, acting via BMI1, promotes an altered cell cycle transition in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2016;304:30–41.
- Chen L, Hu N, Wang C, et al. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle. 2018;17:319–329.
