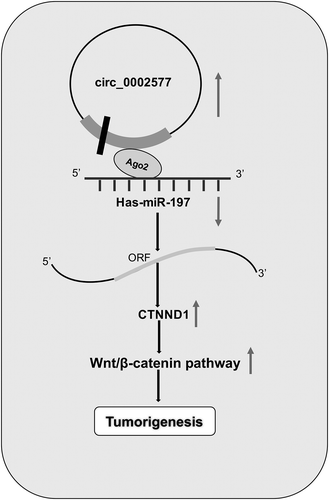
ABSTRACT
Circular RNA (circRNA) is involved in a wide range of life processes including tumorigenesis. However, the molecular mechanisms of circRNA in endometrial carcinoma (EC) carcinogenesis remain unclear. In the present study, we aimed to investigate the potential modulation of hsa_circ_0002577 on EC progression. Here, we showed that hsa_circ_0002577 expression was significantly upregulated in EC tissues, and high hsa_circ_0002577 expression was associated with advanced FIGO stage, lymph node metastasis, and poor overall survival rate of EC patients. In function assays, we demonstrated that hsa_circ_0002577 knockdown significantly reduced EC cells proliferation, migration, invasion ability in vitro and decreased tumor growth in vivo. In mechanism study, we revealed that hsa_circ_0002577 might act as a sponge for miR-197, and CTNND1 was revealed to be a target gene of miR-197. In addition, we revealed that the oncogenic effects of hsa_circ_0002577 were attributed to the regulation of miR-197/CTNND1/Wnt/β-catenin axis. Taken together, we indicated that hsa_circ_0002577 could play critical functions by hsa_circ_0002577/miR-197/CTNND1/Wnt/β-catenin signaling pathway, which served as a novel therapeutic application for EC treatment.
Introduction
Endometrial cancer (EC) is one of the most common malignant neoplasms of the female reproductive tract [Citation1,Citation2]. Strategies for treatment of EC are generally with surgery, and in some cases, a combination of chemotherapy, radiotherapy, and hormone therapy [Citation3]. Although a variety of treatment strategies have been used to improve the survival rate of patients with advanced EC, the clinical outcome is still poor [Citation4]. Therefore, it is urgent to explore the molecules mechanism involved in the pathogenesis of EC.
Circular RNAs (circRNAs), a group of endogenous non-coding RNAs, have been reported to play critical roles at the post-transcriptional level [Citation5,Citation6]. CircRNAs form special closed loop structures lacking the 5ʹ cap and the 3ʹ end of the poly (A) tail and have a higher degree of stability and sequence conservation among mammalian cells [Citation7,Citation8]. Recently, increasing studies explored the roles of circRNAs in the pathogenesis of cancers. For example, Zong et al. found that circRNA_102231 was significantly upregulated in lung cancer patients and associated with advanced TNM stage, lymph node metastasis and poor overall survival [Citation9]. Yang et al. found that silencing of cZNF292 circular RNA suppressed human glioma tube formation via the Wnt/β-catenin signaling pathway [Citation10]. Jin et al. revealed that hsa_circ_0136666 promoted the proliferation and invasion of colorectal cancer through miR‐136/SH2B1 axis [Citation11]. Recently, Xu et al. showed that hsa_circ_0002577 was one of the most upregulated circRNAs in EC samples via circRNA microarray [Citation12]. However, the function and underlying mechanisms of hsa_circ_0002577 are still unclear.
In the present study, we explore the functions of hsa_circ_0002577 in EC progression. Results showed that hsa_circ_0002577 expression was significantly upregulated in EC tissues, and high hsa_circ_0002577 expression was associated with advanced FIGO stage, lymph node metastasis, and poor overall survival rate of EC patients. In function assays, we revealed that hsa_circ_0002577 could promote EC cells proliferation and invasion via miR-197/CTNND1/Wnt/β-catenin axis. Thus, we suggested that hsa_circ_0002577 might act as a potential therapeutic target for EC treatment.
Materials and methods
Patients and clinical tissue samples
A total of 36 EC tissues were collected from EC patients who received hysterectomy at The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology, from January 2014 to December 2016. All samples received no chemotherapy or radiotherapy before surgery. Tissue specimens were immediately stored and kept at −80°C in a refrigerator until RNA extraction. The histological aspects of the samples were judged following the criteria of the International Federation of Gynecology and Obstetrics (FIGO) [Citation13]. Written informed consents were obtained from every participant in this present study, and all the manipulates were approved by the Ethics Committee of Henan University of Science and Technology.
Cell culture and transfection
ECC-1 and HEC-1-A cells were obtained from the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Invitrogen) in a 5% CO2 atmosphere at 37°C.
To block the expression of hsa_circ_0002577 in cells, 3 siRNAs (siRNA1-3) were utilized, and qRT-PCR was used to examine the knockdown efficiency. The hsa_circ_0002577 siRNAs and its negative control (si-NC) were designed and purchased from RiboBio (Guangzhou, China). Cell transfection was completed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions [Citation14].
Quantitative real-time PCR
Total RNA from tissues and cell lines were extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA). RNA was reverse transcribed into cDNA using the GoldenstarTM RT6 cDNA Synthesis Kit (TSINGKE, Beijing, China). QRT-PCR reactions were performed using the QuantStudio Dx Real-Time PCR Instrument (Applied Biosystems, Foster City, CA, USA) and the HieffTM qPCR SYBR® Green Master Mix (YEASEN, Shanghai, China). The qRT-PCR results were normalized to GAPDH and expression fold change was calculated according to the 2−ΔΔCt method.
Western blot
Total proteins were extracted using RIPA lysis buffer (Beyotime, China) and qualified by a BCA kit according to the manufacturer’s instructions. Equal amount protein was separated by dodecyl sulfate, sodium salt (SDS)-Polyacrylamide gel electrophoresis and transferred to PVDF membrane (Millipore, MA, USA). Then, the membrane was blocked with 5% BSA and incubated with primary antibodies (Santa Cruz, CA, USA) at 4°C overnight. Next, the membrane was incubated with the corresponding HRP-conjugated secondary antibodies (Abcam) at room temperature for 1 h [Citation15]. The specific band was determined by ECL chromogenic substrate (Beyotime, China) imaged using a BioSpectrum Imaging System (Bio-Rad, CA, USA).
CCK-8 assay
CCK-8 assay was conducted to detect cell viability using the CCK-8 Kit (Dojindo, Japan). In brief, 2 × 103 cells were seeded into 96-well plates and cultured for different time. Then, 10ul CCK-8 solution was added into the wells and incubated for 2 h at 37℃. The absorbance at 450 nm was measured using a microplate reader (Bio-Rad, CA, USA).
Colony formation assay
Transfected cells were seeded into six-well plates at a concentration of 3000 cells/well and cultured for 2 wk at 37℃ in an incubator with 5% CO2. The colonies were firstly fixed with methanol for at least 20 min and then subjected to stain with Giemsa solution for 15 min. The number of colonies was calculated under a microscope (Nikon, Japan).
Flow cytometry analysis
Transfected cells were digested with trypsin (Thermo Fisher, Waltham, USA), and fixed in 70% ethanol after being washed by ice-cold PBS at 4°C overnight. PI/RNase Staining Buffer (BD Biosciences, San Jose, USA) was used to stain treated cells in cell cycle for 20 min. In addition, the binding buffer containing 5μl PI and 5μl Annexin V-FITC (BD Biosciences) were used to assess apoptosis rate. Cell cycle or apoptosis was observed using the FACS Calibur (BD Biosciences) and analyzed by the use of FlowJo software (FlowJo, Ashland, USA).
Transwell invasion assay
Transwell assay was carried out to measure invasion using the 24-well transwell chambers (8μm pore; Corning, NY, USA) with Matrigel (Corning). In brief, 2 × 104 cells were seeded into the upper chamber in 200μl serum-free medium. The lower chamber was filled with 600μl medium with 10% FBS. After 48 h, the invaded cells on the lower chamber were fixed with 4% formaldehyde and stained with 0.1% crystal violet. Then, cells were counted under digital microscopy (Nikon, Japan).
Luciferase reporter assay
The potential binding sites for miR-197 in hsa_circ_0002577 and CTNND1 were predicted using circinteractome, miRDB, and TargetScan7 tools. The sequences containing these wild-type or mutant binding sites were inserted into the pmirGLO luciferase vector (Promega, Madison, WI, USA) to generate reporter plasmids. For luciferase reporter assay, the reporter plasmid and miR-197 mimics were co-transfected into cells and cultured for 24 h. Then, the luciferase activity was measured using the Dual-luciferase reporter assay system according to the manufacturer’s instructions.
RNA immunoprecipitation
All the steps were performed following the instructions of Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA). Approximately 1 × 107 cells were pelleted and re-suspended with RIP Lysis Buffer plus protease and RNase inhibitors. The 100ul cell lysates were incubated with IgG or antibody at 4°C overnight. The immunoprecipitated RNAs were extracted using the RNeasy MinElute Cleanup Kit (Qiagen) and reverse transcribed using the GoldenstarTM RT6 cDNA Synthesis Kit (TSINGKE, Beijing, China). The enrichment was examined by qRT-PCR assay.
Tumor formation assay in vivo
Four-week-old female BALB/c nude mice were used for the in vivo tumor formation assay. Cells stably transfected with hsa_circ_0002577 shRNAs or its negative control were collected and re-suspended in RPMI-1640 medium at a concentration of 1 × 106 cells/ml, and then 200μl of cell suspension was injected into the right flank of nude mice. The tumor volumes were determined every week, and the volume of tumors was examined as the length×width2 × 0.5. Seven weeks later, the tumor weights were measured. All animal experimental procedures were in agreement with the Ethics Committee of Henan University of Science and Technology.
Statistical analysis
All results were from three independent experiments, analyzed by SPSS 20.0 (SPSS, USA) and expressed as the mean ± SD. The differences were analyzed using the Student’s t-test or one-way ANOVA analysis as appropriate. P < 0.05 was considered a statistically significant difference.
Results
Hsa_circ_0002577 was upregulated in EC
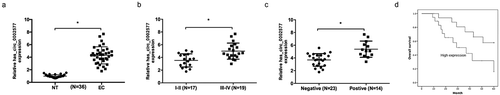
To investigate the roles of hsa_circ_0002577 in EC progression, we determined hsa_circ_0002577 levels in EC tissues, qRT-PCR showed that hsa_circ_0002577 levels were significantly increased in EC tissues ()). And a significant difference was observed between FIGO stage I-II and stage III-IV (). Next, we found that hsa_circ_0002577 levels in patients with lymph node metastases were higher than that in patients without lymph node metastases (). Furthermore, Kaplan-Meier analysis with log-rank test revealed that patients with high hsa_circ_0002577 expression displayed poor overall survival rate ().
Figure 1. Hsa_circ_0002577 was increased in EC. (a) Relative expression of hsa_circ_0002577 in EC tissues was detected by qRT-PCR. (b) Relative expression of hsa_circ_0002577 was increased in EC patients with tumor stage III-IV. (c) relative expression of hsa_circ_0002577 was upregulated in EC patients with lymph node metastases. (d) Kaplan-Meier survival curves showed that high hsa_circ_000257 expression was associated with low overall survival of EC patients. *P < 0.05.
Hsa_circ_0002577 inhibition reduced EC cells proliferation
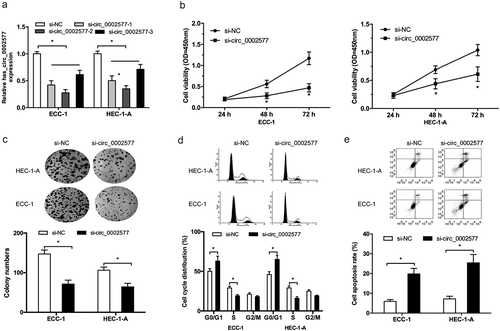
To investigate the roles of hsa_circ_0002577 in EC progression, we decreased hsa_circ_0002577 levels in ECC-1 and HEC-1-A cells (). CCK-8 and Colony formation assays showed that hsa_circ_0002577 inhibition significantly decreased ECC-1 and HEC-1-A cells viability compared to the control group (,)). Furthermore, the effects of hsa_circ_0002577 on EC cells cycle and apoptosis were examined. Flow cytometry analysis showed that hsa_circ_0002577 inhibition arrested ECC-1 and HEC-1-A cells in G0/G1 phase (), and hsa_circ_0002577 suppression induced ECC-1 and HEC-1-A cells apoptosis rate ().
Figure 2. Hsa_circ_0002577 inhibition reduced EC cells proliferation. (a) hsa_circ_0002577 expression was decreased by siRNA transfection in ECC-1 and HEC-1-A cells. (b,c) CCK-8 and colony formation assays were used to explore the effects of hsa_circ_0002577 on EC cells proliferation. (d,e) flow cytometry analysis was used to determine the effects of hsa_circ_0002577 on EC cells cycle and apoptosis. *P < 0.05.
Hsa_circ_0002577 inhibition suppressed EC cells invasion
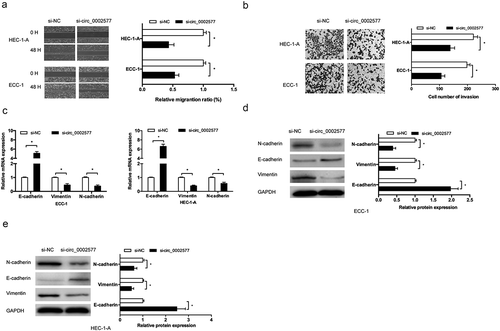
Next, we explored the effects of hsa_circ_0002577 on EC cells migration and invasion ability. Wound healing assay showed that hsa_circ_0002577 suppression drastically attenuated cellular migration ability in ECC-1 and HEC-1-A cells (). Meanwhile, Transwell invasion assay suggested that knockdown of hsa_circ_0002577 reduced the invasive ability of ECC-1 and HEC-1-A cells (). Moreover, the expression of epithelial–mesenchymal transition (EMT) markers such as E-cadherin, N-cadherin, and Vimentin in EC cells was further determined. Results showed that hsa_circ_0002577 suppression decreased the levels of N-cadherin and Vimentin while increased the levels of E-cadherin both in mRNA () and protein (, )) levels.
Figure 3. Hsa_circ_0002577 inhibition reduced EC cells invasion and EMT processes. (a,b) wound healing assay and transwell invasion assay were used to determine the effects of hsa_circ_0002577 on EC cells migration and invasion ability. (c,d) QRT-PCR and western blot assays were used to explore the effects of hsa_circ_0002577 on EMT markers (E-cadherin, N-cadherin, and vimentin) in ECC-1 and HEC-1-A cells. *P < 0.05.
Hsa_circ_0002577 acted as a sponge for mir-197
Presently, circRNAs was widely considered as a miRNA “sponge” and may involve in cancer occurrence and development [Citation16]. To further investigate the underlying molecular mechanisms of hsa_circ_0002577 in EC progression, we screened the target miRNAs of hsa_circ_0002577 by bioinformatics analysis. We found that hsa_circ_0002577 might interact with several miRNAs, the qRT-PCR analysis revealed that miR-197 had the highest enrichment value compared with other miRNAs (), indicating miR-197 might be a direct target of hsa_circ_0002577 in EC (). Dual luciferase reporter assay showed that miR-197 mimics significantly reduced the luciferase activity of circ_0002577 Wt in ECC-1 and HEC-1-A cells (). RNA precipitation (RIP) results showed that hsa_circ_0002577 and miR-197 were preferentially enriched in Ago2-containing miRNPs compared to anti-IgG immunoprecipitates (). In addition, we explored miR-197 expression in EC tissues, results showed that miR-197 expression was downregulated and negatively correlated with hsa_circ_0002577 levels in EC patients (, )), which was further confirmed in starbase database ().
Figure 4. Hsa_circ_0002577 functioned as a molecular sponge for miR-197. (a) QRT-PCR revealed that miR-197 had the highest enrichment value compared with other miRNAs. (b) The predicted binding sites for miR-197 in the 3ʹUTR of hsa_circ_0002577. (c) Dual luciferase reporter assay showed that miR-197 mimics significantly reduced the luciferase activity of circ_0002577 Wt in EC cells. (d) RIP assay showed that hsa_circ_0002577 and miR-197 were enriched in Ago2-containing miRNPs. (e) Relative expression of miR-197 was decreased in EC tissues. (f) MiR-197 expression was negatively correlated with hsa_circ_0002577 expression in EC tissues. (g) Relative expression of miR-197 in EC tissues was detected by starbase database. (h,i) CCK-8 and transwell invasion assays showed that miR-197 inhibitors reversed the effects of hsa_circ_0002577 inhibition on EC cells proliferation and invasion. *P < 0.05.
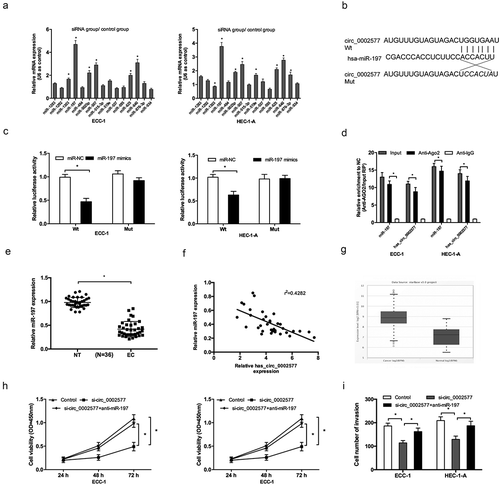
To further determine whether miR-197 inhibition is the functional mechanism of hsa_circ_0002577, we transduced si-circ_0002577 and miR-197 inhibitors (anti-miR-197) in EC cells. In vitro functional assays showed that miR-197 inhibitors significantly reversed the effects of hsa_circ_0002577 inhibition on EC cells proliferation and invasion ability (, )).
CTNND1 acted as a direct target of mir-197
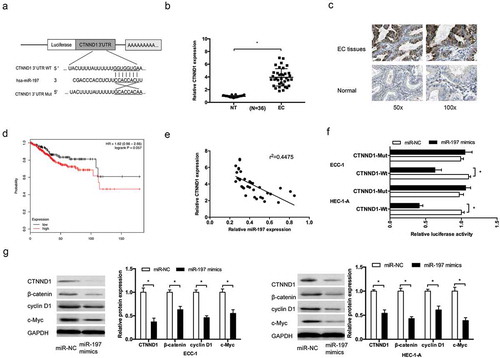
We utilized computational prediction TargetScan to identify the potential binding sites for CTNND1 mRNA on miR-197. We found the 3ʹUTR of CTNND1 mRNA have potential miR-197 binding site (). Next, we explored CTNND1 expression in EC tissues. QRT-PCR showed that CTNND1 expression was upregulated in EC tissues (), which was also further confirmed by IHC in EC tissues (). Kaplan–Meier Plotter analysis showed that high CTNND1 expression was associated with poor overall survival of EC patients (). Correlation analysis showed that miR-197 was negatively correlated with CTNND1 expression in EC tissues ()). Dual luciferase reporter assay showed that miR-197 mimics reduced the luciferase activity of CTNND1-Wt in EC cells (). Furthermore, Western blot showed that miR-197 mimics significantly suppressed CTNND1, β-catenin, cyclin D1, and c-Myc expression in EC cells (). Therefore, CTNND1 might be a direct target of miR-197, and miR-197 might affect the activity of CTNND1/Wnt/β-catenin pathway in EC progression.
Figure 5. CTNND1 was a direct target of miR-197. (a) Potential binding site for miR-197 in the 3ʹUTR of CTNND1. (b,c) QRT-PCR and IHC were used to explore CTNND1 expression in EC tissues. (d) Kaplan-Meier plotter analysis showed that high CTNND1 expression was associated with poor overall survival of EC patients. (e) MiR-197 expression was negatively correlated with hsa_circ_0002577 expression in EC tissues. (f) Dual luciferase reporter assay showed that miR-197 mimics reduced the luciferase activity of CTNND1-Wt in EC cells. (g) Western blot showed that miR-197 mimics significantly suppressed CTNND1, β-catenin, cyclin D1, and c-Myc expression in EC cells. *P < 0.05.
Hsa_circ_0002577 regulated ctnnd1/wnt/β-catenin pathway and EC progression via mir-197
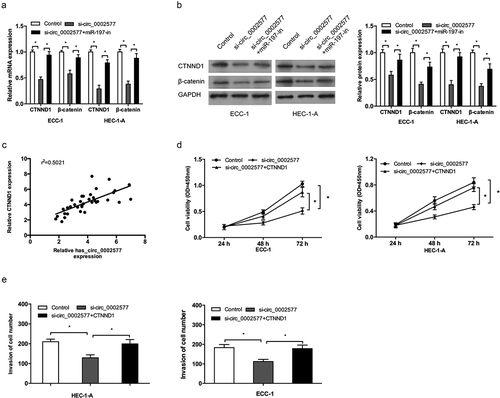
As the above paragraph speculated the relationship between hsa_circ_0002577, miR-197, and CTNND1. Next, we confirmed whether hsa_circ_0002577 could regulate CTNND1/Wnt/β-catenin via miR-197. QRT-PCR and Western bolt showed that hsa_circ_0002577 inhibition significantly downregulated CTNND1 and β-catenin expression, while the effects induced by hsa_circ_0002577 inhibition could be abolished by miR-197 inhibitors (, )). Next, correlation analysis revealed that there is a positive correlation between hsa_circ_0002577 and CTNND1 expression in EC tissues (). To further determine the roles of hsa_circ_0002577/miR-197/CTNND1 axis in EC progression, EC cells were transfected with si-circ_0002577 or si-circ_0002577 + CTNND1 plasmid. CCK-8 and transwell invasion assays showed that hsa_circ_0002577 knockdown decreased the proliferation and invasion of EC cells, which could be abolished by CTNND1 plasmid transfections (, )).
Figure 6. Restoration of CTNND1 abrogated the effects of hsa_circ_0002577 knockdown. (a,b) QRT-PCR and western bolt showed that hsa_circ_0002577 inhibition decreased CTNND1, and β-catenin expression, which could be abolished by miR-197 inhibitors. (c) CTNND1 expression was positively correlated with hsa_circ_0002577 expression in EC tissues. (d,e) CCK-8 and transwell invasion assays showed that hsa_circ_0002577 knockdown decreased the proliferation and invasion ability of EC cells, which could be abolished by CTNND1 overexpression. *P < 0.05.
Hsa_circ_0002577 knockdown reduced tumor growth in vivo
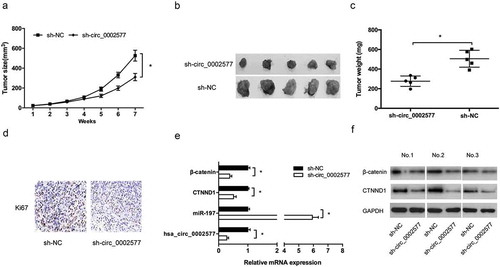
Next, we explored the effects of hsa_circ_0002577 on EC growth in vivo. sh-circ_0002577 transfected ECC-1 cells were injected into the flank of nude mice. We found that hsa_circ_0002577 knockdown significantly decreased tumor volumes (). At 7 wk after injection, the mice were sacrificed and tumor weights were determined. Results showed that hsa_circ_0002577 suppression significantly reduced tumor weights (, )). Moreover, hsa_circ_0002577 inhibition xenograft tumors displayed decreased Ki-67 expression compared to the control group (). Next, we explored hsa_circ_0002577, miR-197, CTNND1 and β-catenin expression in xenograft tumors, Results showed that sh-circ_0002577 significantly reduced hsa_circ_0002577, CTNND1, and β-catenin expression, while increased miR-197 expression in xenograft tumors (, )). Thus, we suggested that hsa_circ_0002577 could promote EC progression via miR-197/CTNND1/Wnt/β-catenin axis ().
Figure 7. Hsa_circ_0002577 inhibition suppressed EC growth in vivo. (a,b) Effect of sh-circ_0002577 on tumor growth. (c) Tumor weight of the sh-circ_0002577 group was significantly reduced than that of the sh-NC group. (d) Ki67 expression was determined by IHC in xenograft tumors. (e) Hsa_circ_0002577, miR-197, CTNND1 and β-catenin expression in xenograft tumors were determined by qRT-PCR. (f) CTNND1 and β-catenin protein expression in xenograft tumors were determined by Western blot. *P < 0.05.
Discussion
CircRNAs were thought to be the products of transcription errors earlier and until 2012, along with the development of high-throughput sequencing technologies and bioinformatics, Salzman made a comprehensive and systematic report on circRNAs and their general features in the gene expression program in human cells, making it a hotspot in non-coding RNA fields [Citation17]. Recent studies showed that lots of circRNAs were dysregulated in gynecology cancers, which may contribute to tumor abnormal growth and metastasis. For example, Liu et al. showed that circRNA8924 promoted cervical cancer cell proliferation, migration, and invasion by competitively binding to miR-518d-5p/519-5p family and modulating the expression of CBX8 [Citation18]. Hu et al. showed that circular RNA circ-ITCH suppressed ovarian carcinoma progression through targeting miR-145/RASA1 signaling [Citation19]. Chen et al. found that hsa_circ_0061140 knockdown reversed foxm1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity [Citation20].
In the present study, we showed that hsa_circ_0002577 was increased and associated with advanced tumor stage, lymph node metastases, and poor overall survival rate of EC patients. However, the number of EC specimens is small, more EC specimens will be explored in our further study. Functional assays revealed that hsa_circ_0002577 significantly increased EC cells proliferation, invasion, and EMT processes. In addition, we found that hsa_circ_0002577 knockdown reduced tumor growth in vivo. These data demonstrated that hsa_circ_0002577 might act as a critical tumor oncogenic circRNA in EC tumorigenesis.
Accumulating evidence suggested that circRNAs play critical roles in regulating gene expression by sequestering the miRNAs [Citation21]. miRNAs could regulate oncogenes or tumor suppressor genes to mediate cell metastasis, metabolism, and proliferation in cancer [Citation22,Citation23]. Previous studies showed that miR-197 plays important roles in tumor progression. For example, Hu et al. found that miR-197 was downregulated in cervical cancer and suppressed cell proliferation and invasion through targeting forkhead box M1 [Citation24]. Wu et al. revealed that lncRNA SNHG20 promoted the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis [Citation25]. However, the expression and underlying mechanisms of miR-197 in EC progression remain unclear. In the present study, miR-197 was confirmed as a novel interactive miRNA of hsa_circ_0002577. MiR-197 expression was decreased and negatively associated with hsa_circ_0002577 expression in EC tissues. Furthermore, in vitro function assays, we showed that miR-197 inhibitors significantly reversed the effects of hsa_circ_0002577 knockdown on EC cells’ progression, indicating miR-197 might act as a tumor suppressor in EC progression.
Catenin delta 1 (CTNND1), also known as p120-catenin, was originally identified as a substrate of the oncogenic tyrosine kinase Src [Citation26] and subsequently defined as a component of the adherens junction complex that includes E-cadherin and α-, β-, γ-catenins [Citation27]. Recent studies showed that CTNND1 could act as a promising therapeutic target for cancer treatment. For example, Cao et al. showed that microRNA-298 repressed hepatocellular carcinoma progression by inhibiting CTNND1-mediated Wnt/β-catenin signaling [Citation28]. Tang et al. found that overexpression of CTNND1 in hepatocellular carcinoma promoted tumor progression through activation of Wnt/β-catenin signaling [Citation29]. In our study, CTNND1 was identified as a direct target of miR-197, and miR-197 might affect the activity of CTNND1/Wnt/β-catenin pathway in EC progression. Furthermore, we showed a positive correlation between hsa_circ_0002577 expression and CTNND1 in EC tissues. Hsa_circ_0002577 inhibition downregulated CTNND1, β-catenin, cyclin D1, and c-Myc expression, while the effects induced by hsa_circ_0002577 inhibition could be abolished by miR-197 inhibitors. In addition, we showed that hsa_circ_0002577 knockdown decreased the proliferation and invasion of EC cells, which could be abolished by CTNND1 plasmid transfections. Therefore, the present study demonstrated that hsa_circ_0002577/miR-197/CTNND1/Wnt/β-catenin pathway could play important roles in EC progression.
In conclusion, the present study revealed a significant role of a novel circRNA hsa_circ_0002577 in EC progression and suggested that hsa_circ_0002577/miR-197/CTNND1/Wnt/β-catenin pathway might be a potential therapeutic for EC treatment.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
- Purdie DM, Green AC. Epidemiology of endometrial cancer. Best Pract Res Clin Obstetrics Gynaecol. 2001;15(3):341–354.
- Hüsing A, Dossus L, Ferrari P, et al. An epidemiological model for prediction of endometrial cancer risk in Europe. Eur J Epidemiol. 2016;31(1):51–60.
- Fader AN, Arriba LN, Frasure HE, et al. Endometrial cancer and obesity: epidemiology, biomarkers, prevention and survivorship. Gynecol Oncol. 2009;114(1):121–127.
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157.
- Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20(12):1829–1842.
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205.
- Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. 2018;102:639–644.
- Yang P, Qiu Z, Jiang Y, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/β-catenin signaling pathway. Oncotarget. 2016;7(39):63449.
- Jin C, Wang A, Liu L, et al. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR‐136/SH2B1 axis. J Cell Physiol. 2019;234(5):7247–7256.
- Xu H, Gong Z, Shen Y, et al. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics. 2018;10(2):187–197.
- Di Renzo GC, Conry JA, Blake J, et al. International Federation of Gynecology and Obstetrics opinion on reproductive health impacts of exposure to toxic environmental chemicals. Int J Gynecol Obstet. 2015;131(3):219–225.
- Zhang H, Yang F, Chen SJ, et al. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumor Biol. 2015;36(4):2947–2955.
- Yang F, Zhang H, Chen SJ, et al. MiR-506 is down-regulated in clear cell renal cell carcinoma and inhibits cell growth and metastasis via targeting FLOT1. PloS One. 2015;10(3):e0120258.
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521.
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012;7(2):e30733.
- Liu J, Wang D, Long Z, et al. CircRNA8924 promotes cervical Cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p/519-5p family and modulating the expression of CBX8. Cell Physiol Biochem. 2018;48(1):173–184.
- Hu J, Wang L, Chen J, et al. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem Biophys Res Commun. 2018;505(1):222–228.
- Chen Q, Zhang J, He Y, et al. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55–63.
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148.
- Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metast Rev. 2009;28(3–4):369.
- Farazi TA, Spitzer JI, Morozov P, et al. miRNAs in human cancer. J Pathol. 2011;223(2):102–115.
- Hu Q, Du K, Mao X, et al. miR-197 is downregulated in cervical carcinogenesis and suppresses cell proliferation and invasion through targeting forkhead box M1. Oncol Lett. 2018;15(6):10063–10069.
- Wu J, Zhao W, Wang Z, et al. Long non‐coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR‐197/LIN28 axis. J Cell Mol Med. 2019;23(1):680–688.
- Reynolds AB, Roesel DJ, Kanner SB, et al. Transformation-specific tyrosine phosphorylation of a novel cellular protein in chicken cells expressing oncogenic variants of the avian cellular src gene. Mol Cell Biol. 1989;9(2):629–638.
- Shibamoto S, Hayakawa M, Takeuchi K, et al. Association of p120, a tyrosine kinase substrate, with E-cadherin/catenin complexes. J Cell Biol. 1995;128(5):949–957.
- Cao N, Mu L, Yang W, et al. MicroRNA-298 represses hepatocellular carcinoma progression by inhibiting CTNND1-mediated Wnt/β-catenin signaling. Biomed Pharmacother. 2018;106:483–490.
- Tang B, Tang F, Wang Z, et al. Overexpression of CTNND1 in hepatocellular carcinoma promotes carcinous characters through activation of Wnt/β-catenin signaling. J Exp Clin Cancer Res. 2016;35(1):82.