ABSTRACT
During mitosis, Aurora B kinase is required for forming proper bi-oriented kinetochore-microtubule attachments. Current models suggest that tension exerted between a pair of sister-kinetochores (inter-kinetochore stretch) produces a spatial separation of Aurora B kinase from kinetochore-associated microtubule binding substrates, such as the Knl1-Mis12-Ndc80 (KMN) network, resulting in a decrease of phosphorylation and, thus, an increase of affinity for microtubules. Using Single-Molecule High-Resolution Colocalization (SHREC) microscopy analysis of the kinetochore-associated motor CENP-E, we now show that CENP-E undergoes structural rearrangements prior to and after tension generation at the kinetochore, and displays a bi-modal Gaussian distribution on a pair of bi-oriented sister kinetochores. The conformational change of CENP-E depends on its microtubule-stimulated motor motility and the highly flexible coiled-coil between its motor and kinetochore-binding tail domains. Chemical inhibition of the motor motility or perturbations of the coiled-coil domain of CENP-E increases Aurora B-mediated Ndc80 phosphorylation in a tension-independent manner. Metaphase chromosome misalignment caused by CENP-E inhibition can be rescued by chemical inhibition of Aurora B kinase. Furthermore, a pair of monotelic sister-kinetochores shows asymmetric levels of Aurora B-mediated phosphorylation in mono-polar spindles depending on CENP-E motor activity. These results collectively suggest a tension-independent mechanism to reduce Aurora B-mediated phosphorylation of outer kinetochore components in response to microtubule capture by CENP-E.
Introduction
The major task to ensure accurate chromosome segregation is to stabilize proper bi-oriented attachments, in which one sister kinetochore captures microtubules from one spindle pole and the other from the opposite pole, and resolve improper attachments, such as syntelic attachments (both sister kinetochores bind to microtubules from the same pole). Aurora B kinase is a key component involved in the attachment error correction process [Citation1,Citation2]. The current “spatial separation” model [Citation1] suggests that tension exerted between sister kinetochores with correct end-on attachments results in a spatial separation of the inner centromere-associated Aurora B from its substrates localized at the outer kinetochore. This arrangement would minimize Aurora B-mediated phosphorylation of the core microtubule binding proteins, e.g. Ndc80, at kinetochores with proper microtubule attachments. In contrast, high levels of phosphorylation would persist at kinetochores with incorrect attachments. This prevailing model relies on the distance between the inner-centromere-localized Aurora B kinase and the outer-kinetochore-localized phosphatases as being critical for the balance of phosphorylation/dephosphorylation. Quantitatively measuring the fraction of Ndc80 complexes bound to microtubules at individual kinetochores in living human cells has shown that Ndc80 binding is predominantly regulated by centromere tension and the proper localization of Aurora B kinase [Citation3]. In addition, protein phosphatases, including PP1 and PP2A, have been shown to be recruited to kinetochores reversing the phosphorylation of core microtubule binding proteins and promoting microtubule attachment [Citation4].
As a kinetochore-associated plus-end directed motor, CENP-E plays a critical role in ensuring cells to segregate their chromosomes accurately. CENP-E has been proposed to power chromosome movement along spindle microtubules [Citation5] and/or to maintain a mechanical link between kinetochores and the plus-end tips of dynamic microtubules [Citation6,Citation7]. Chemical inhibition of CENP-E motor activity, removal of CENP-E from cells, or mutational analysis in mice leads to an obvious metaphase plate with a few unaligned chromosomes [Citation8–Citation11]. Furthermore, the CENP-E motor activity has been shown to be regulated by an Aurora kinases/PP1 phosphorylation switch [Citation12].
Structural studies of CENP-E have revealed that unlike conventional kinesin motors, which have a short coiled-coil domain either rigidly extended or folded through hinge segments in the middle, CENP-E has a discontinuous coiled-coil domain that is approximately 230 nm long [Citation13]. This unique coiled-coil enables CENP-E to rearrange into different conformations in vitro [Citation13]. We now find that kinetochore-associated CENP-E undergoes structural changes that are responsive to microtubule capture and microtubule dynamics, and displays a bi-modal Gaussian distribution at a pair of bi-oriented sister kinetochores. Microtubule capture by motor CENP-E at kinetochores triggers a tension-independent mechanism reducing Aurora B-mediated phosphorylation of the Ndc80 complex and, thus, enhancing connections between kinetochores and dynamic microtubule plus ends.
Results
The long and flexible coiled-coil domain of CENP-E is essential for the kinetochore function of CENP-E
Unlike conventional kinesins, CENP-E has a long and flexible coiled-coil domain between its N-terminal motor domain and C-terminal kinetochore-binding domain [Citation13]. To test the role of this long and flexible coiled-coil domain, we designed several CENP-E constructs tagged with GFP at the carboxyl-termini ()), including a Full-length-CENP-E construct as a control and a Tail-CENP-E construct that lacks the motor domain along with a substantial part of the coiled-coil domain. A Mini-CENP-E construct was generated by combining the motor, the tail and a shorter segment of the coiled-coil domain of CENP-E. Furthermore, a part of the flexible coiled-coil of CENP-E was replaced with a more rigid coiled-coil domain from a Kinesin-1 family member, Kif5B, to construct a Chimera-CENP-E. Both Mini- and Chimera-CENP-E retain the Aurora B phosphorylation sites/PP1-docking motif. Cells were co-transfected with CENP-E constructs along with a siRNA that binds to the 3ʹ untranslated region of CENP-E mRNA targeting CENP-E for degradation. Immunoblotting analysis (Figure S1, a and b) and indirect immunofluorescence analysis (Figure S1, c–e) showed a significant reduction of CENP-E protein levels and levels of CENP-E associated with kinetochores, respectively, in cells transfected with CENP-E siRNA compared to cells transfected with control siRNA (Mock). Furthermore, all CENP-E constructs associated with kinetochores at a similar level to unattached (subjected to nocodazole treatment), aligned and unaligned kinetochores as Full-length-CENP-E (Figure S2).
Figure 1. The long and highly flexible coiled-coil domain of CENP-E is essential for its kinetochore function. (a) Illustration (not drawn to scale) of GFP-tagged CENP-E constructs generated in this study. (b) Mini- and Chimera-CENP-E cannot rescue chromosome alignment defects caused by depletion of endogenous CENP-E. Immunofluorescence images were acquired using antibodies against centromere (ACA, a centromere/kinetochore marker, in upper panels) and tubulin (microtubules in lower panels) in human T98G cells. DNA was visualized by DAPI in lower panels. The percentage (mean ± SD) of mitotic cells with fully aligned chromosomes from three independent experiments are summarized (Mock siRNA: n = 644, CENP-E siRNA: n = 680, FL: n = 699, Tail: n = 621, Mini: n = 458, Chimera: n = 614, and CENP-E inhibitor GSK923295 (GSK) n = 686 cells). Scale bar, 5 μm. (c) Expressing Tail-, Mini-, and Chimera-CENP-E as well as inhibition of CENP-E motor activity have dominant negative effects on chromosome misalignment compared to CENP-E depletion. Histogram showing the number of polar chromosomes per metaphase cell (mean ± SD from three independent experiments) in cells co-transfected with the CENP-E siRNA and CENP-E constructs as indicated (CENP-E siRNA: n = 272, Tail: n = 376, Mini: n = 186, Chimera: n = 324, and GSK: n = 404). Two-way ANOVA followed by a Tukey’s multiple comparison test was used to compare the means (**** p < 0.0001). (d) The long and flexible coiled-coil is required for CENP-E motor activity. Immunofluorescence images of monastrol-treated cells expressing CENP-E constructs in cells depleted of endogenous CENP-E or treated with GSK923295 as indicated. Spindle poles and kinetochores were stained using antibodies against γ-tubulin and ACA, respectively. Scale bar, 5 μm. (e) Quantification (Mean ± SD from three independent experiments) of the distance of kinetochores from centrosomes shown in (d) (n = 30 cells per each condition). An unpaired t test or a One-way ANOVA followed by a Sidak’s multiple comparison test was used to compare the means (****p < 0.0001).
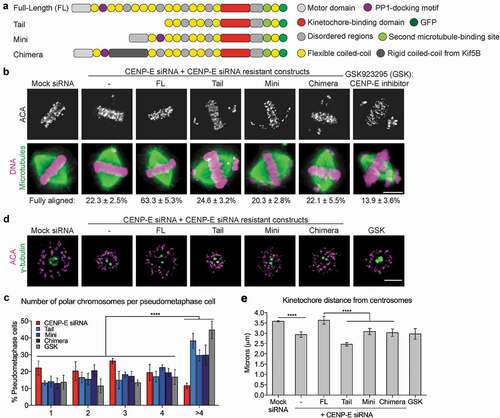
We assessed the chromosome alignment phenotype by indirect immunofluorescence. Cells were partly synchronized using monastrol treatment and then released into MG132 prior to fixation. Consistent with what has been shown before [Citation9,Citation14], the majority of cells depleted of CENP-E showed an obvious metaphase plate with a few polar chromosomes. Expressing only Full-length-CENP-E, but not Tail-, Mini- or Chimera-CENP-E, rescued the chromosome alignment defect ()). More detailed analysis of the number of polar chromosomes per cell showed that most of CENP-E-depleted cells (~80%) had only 1–4 polar chromosomes, whereas expressing Tail-, Mini- or Chimera-CENP-E led to a ~ 20–40% increase in cells with more than 4 polar chromosomes ()). Further, cells treated with GSK923295, an ATPase antagonist of CENP-E that inhibits the motor motility [Citation15], resulted in a similar increase of misaligned chromosome number as CENP-E mutants ()). These results suggest that expressing a motorless, a shorter or a less flexible CENP-E, or chemical inhibition of CENP-E motility not only cannot rescue the chromosome alignment defects caused by CENP-E depletion but also exacerbates the misalignment phenotype characteristic of CENP-E-depleted cells.
It has been shown that the coiled-coil domain of CENP-E regulates the motor function of CENP-E in vitro [Citation16]. To directly test whether Mini- or Chimera-CENP-E has defective motility in cells, we measured chromosome ejection from mono-poles upon inhibition of Kinesin-5 [Citation17]. Depletion of CENP-E or inhibition of CENP-E motility with GSK923295 resulted in reduced distances between kinetochores and spindle poles ()), consistent with the previous finding that CENP-E at kinetochores drives congression of polar chromosomes [Citation18]. Only Full-length-CENP-E, but not Mini- or Chimera-CENP-E, was able to completely rescue the chromosome ejection defect caused by CENP-E depletion ()). These results support the notion that both the length and the flexibility of the coiled-coil are required for the motility of kinetochore-associated CENP-E.
The long and flexible coiled-coil domain of CENP-E regulates the structural flexibility of kinetochore-associated CENP-E
To investigate whether the coiled-coil domain of CENP-E regulates the structural flexibility of CENP-E at the kinetochore with sub-pixel accuracy, we sub-cloned the Full-length-, Mini- and Chimera-CENP-E constructs into a double-tagged, mCherry and EmeraldGFP, vector ()). Single Molecule High-Resolution Co-localization (SHREC) microscopy [Citation19] was adopted to measure the intra-molecular distance between the motor domain and the tail domain of kinetochore-associated CENP-E below the diffraction limit of the light microscope. This super-resolution technique allowed us to obtain the 3-dimensional centroid position of kinetochore-associated CENP-E based on the Gaussian distribution of mCherry and EmeraldGFP fluorescence signals.
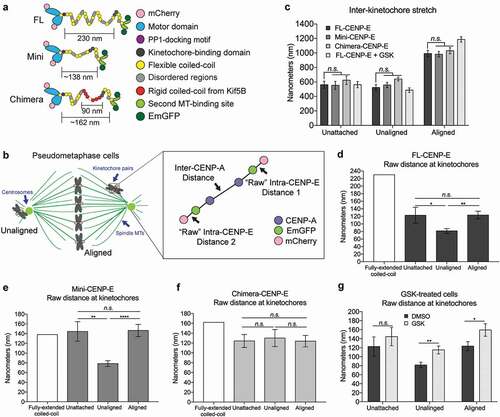
Figure 2. Differential features of CENP-E’s coiled-coil domain and motor domain activity mediate the structural rearrangement of CENP-E. (a) Illustration (not drawn to scale) of transgenes used for CENP-E structural rearrangement analysis at the kinetochore. (b) Illustration (not drawn to scale) of pseudo-metaphase cells depicting unaligned and aligned kinetochores used to analyze the inter-kinetochore distance of CENP-A and intra-kinetochore distance between CENP-E motor and tail domains. (c) Inhibition of CENP-E motility does not affect inter-kinetochore stretch (tension). Quantification (mean ± SD of three independent experiments) of the inter-CENP-A distance at unattached kinetochores in nocodazole treated cells and bi-oriented kinetochores (aligned) and unaligned kinetochores in pseudo-metaphase cells (Unattached, FL: n = 15, Mini: n = 15, and Chimera: n = 16; Unaligned DMSO, FL: n = 36, Mini: n = 33, Chimera: n = 32, and GSK: n = 33; and Aligned DMSO, FL: n = 33, Mini: n = 30, Chimera n = 33; and GSK: n = 36). Two-way ANOVA followed by a Tukey’s multiple comparisons test was used to compare the means (n.s., not significant). (d-f) Both the length and the flexibility of the coiled-coil domain affect CENP-E conformation at the kinetochore in response to different microtubule attachment status. Bar graph (mean ± SEM of three independent experiments) showing the predicted distance (white) along with the measured distance between N-terminal mCherry and C-terminal EmGFP tags (intra-CENP-E) on a single kinetochore in cells expressing (d) FL-, (e) Mini- and (f) Chimera-CENP-E constructs as indicated. In (d), unattached: n = 30, unaligned: n = 72, and aligned: n = 66. In (e), unattached: n = 30, unaligned: n = 66, and aligned: n = 60. In (f), unattached: n = 32, unaligned: n = 66, and aligned: n = 66. One-way ANOVA followed by a Tukey’s multiple comparisons test was used to compare the means (* p < 0.05, ** p < 0.01, **** p < 0.0001, and n.s., not significant). (g) CENP-E motor activity triggers CENP-E conformational changes upon microtubules capture. Bar graph (mean ± SEM of three independent experiments) showing the distance between CENP-E motor domain and tail domain at unattached, unaligned and aligned kinetochores of cells treated with DMSO or GSK923295 (GSK) (Unattached kinetochores, DMSO: n = 30 and GSK: n = 32; unaligned kinetochores, DMSO: n = 72 and GSK: n = 78; and aligned kinetochores, DMSO: n = 66 and GSK: n = 72). An unpaired t test was used to compare the means (* p < 0.05, ** p < 0.01, and n.s., not significant).
We validated our SHREC imaging method by measuring the distance of the outer-kinetochore component, Hec1 (a component of the Ndc80 complex), from the inner kinetochore marker, YFP-CENP-A (Figure S3a). After kinetochore tilt correction using the 3D-SHREC imaging method, we found a similar length of Hec1 relative to YFP-CENP-A (Figure S3b) as previously measured [Citation20]. Nonetheless, because the “delta method” did not allow us to measure the intra-molecular distance of CENP-E directly, we used the “raw distance method” as previously described [Citation21]. The “raw distance method” was also validated by measuring the distance between Hec1 and YFP-CENP-A after chromatic shift correction as previously measured [Citation21]. Using this method, we also found a similar distance of Hec1 relative to YFP-CENP-A (Figure S3C). Therefore, we proceeded to the intra-CENP-E distance analysis with the “raw distance method”.
To determine whether microtubule capture by kinetochores and/or subsequent kinetochore bi-orientation affect the structural behaviour of CENP-E, we specifically chose and analyzed pseudo-metaphase cells with aligned kinetochores at the metaphase plate and a few unaligned kinetochores near spindle poles, including cells expressing Full-length CENP-E ()). We assessed the microtubule attachment status of aligned and unaligned kinetochores using indirect immunofluorescence analysis of Mad1, a microtubule attachment marker [Citation22]. Cells expressing Full-length-, Mini- or Chimera-CENP-E all showed little to no fluorescence signal of Mad1 at aligned kinetochores, indicative of the presence of kinetochore-microtubule attachments; in contrast, the unaligned kinetochores showed high levels of Mad1 fluorescence signals (Figure S4). Sister kinetochore bi-orientation upon proper microtubule attachment for aligned kinetochores at the metaphase plate was further confirmed by measuring the distance between the kinetochore marker, CENP-A, in sister kinetochore pairs. The increased inter-kinetochore stretch at aligned kinetochores compared to the distance at unaligned sister kinetochores confirmed bi-orientation ()). Taken together, we conclude that aligned kinetochores indeed represent pairs of bi-oriented end-on attached kinetochores, whereas unaligned kinetochores are comprised of laterally attached and unattached kinetochores.
To establish the “resting” distance of the intra-molecular distance of CENP-E, we analyzed kinetochores that lack microtubule attachments. To produce unattached kinetochores, we treated cells with nocodazole to depolymerize microtubules. Short-term treatment with nocodazole (15 min) depolymerized all microtubules (Figure S5a), but prevented the kinetochore protein expansion that occurs after prolonged nocodazole treatment (Figure S5b), as previously reported [Citation23]. The distance between sister kinetochore pairs (inter-CENP-A) was measured at approximately 0.5 µm as previously reported [Citation24] and was not affected upon expression of Mini- or Chimera-CENP-E compared to cells expressing Full-length-CENP-E ()). The predicted length of a fully-extended CENP-E molecule was previously measured at approximately 230 nm in vitro by electron microscopy [Citation13]. On unattached kinetochores, the intra-molecular distance of Full-length-CENP-E was halved of the predicted fully-extended length, averaging at 122.8 nm ()). In contrast, Mini-CENP-E had an average of 144.6 nm intra-molecular distance ()), in which the majority of the measured distances were close to the predicted length of approximately 138 nm, suggesting a fully-extended configuration of Mini-CENP-E. Replacing the flexible coiled-coil domain with Kif5B’s coiled-coil (Chimera-CENP-E) resulted in an almost fully-extended configuration, a predicted length of approximately 162 nm vs. the measured distance of 124.2 nm ()). Therefore, the length and the flexibility of the coiled-coil domain of CENP-E is important for a folded configuration of kinetochore-associated CENP-E in the absence of microtubule attachments.
Next, we analyzed the intra-CENP-E distance in pseudo-metaphase cells on both unaligned and aligned kinetochores (Figure S5, c and d). Full-length-CENP-E at unaligned kinetochores showed a further decrease in the intra-molecular distance of CNEP-E compared to unattached kinetochores, from 122.8 nm at unattached kinetochores to 81.5 nm at unaligned kinetochores ()). The Mini-CENP-E showed a decrease in the intra-molecular distance that was similar to the Full-length-CENP-E control at unaligned kinetochores ()). Conversely, the Chimera-CENP-E mutant remained in an extended configuration in all conditions analyzed at approximately 120 nm at unattached, unaligned and aligned kinetochores ()). These results indicate that force generated by the motor activity upon microtubule capture by CENP-E on unaligned kinetochores driving them to the metaphase plate produces a conformational change and this change depends on the super flexible coiled-coil of CENP-E. To confirm that the motor motility of CENP-E plays a role in regulating the structural flexibility of CENP-E upon microtubule capture, we treated cells with GSK923295. Indeed, motility inhibition of Full-length-CENP-E construct resulted in a significant increase in the intra-molecular distance at both aligned and unaligned kinetochores, but not unattached kinetochores ()).
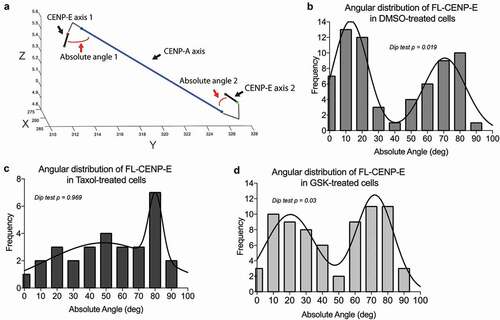
Analysis of the intra-molecular distance of Mini-CENP-E ()) showed a similar pattern at unaligned and aligned kinetochores compared to Full-length-CENP-E ()). This was unexpected given that Mini-CENP-E has a reduced coiled-coil length as well as a defective motor motility similar to Chimera-CENP-E ()). However, an average length between the motor domain and the tail domain could represent either one homogenous population of CENP-E at kinetochores or a heterogeneous population with some elongated molecules and some shortened ones (Figure S6). Therefore, we conducted further structural analyses of kinetochore-associated CENP-E by examining the angular distribution of CENP-E relative to the kinetochore axis (inter-CENP-A) in a 3-dimensional space () and Supplemental Movie S1). The angles measured in these analyses are between the CENP-A axis connecting two sister kinetochores and the specific axis connecting N- and C-terminal fluorophores of CENP-E molecules on a population scale. Angular analysis of Full-length-CENP-E at bi-oriented kinetochores revealed a two-population cluster that followed a bi-modal Gaussian distribution with local maxima at approximately 14° and 68° ()). We did not find this bi-modal Gaussian distribution in cells expressing Mini-CENP-E (Figures S7). Density plotting of angular distributions of cells treated with Taxol, which disrupts microtubule dynamics, affected the bi-modal Gaussian distribution of Full-length CENP-E causing a shift towards a local maximum of 72° ()). In contrast, a short-term treatment of GSK-923295, a specific allosteric inhibitor of CENP-E that could lock CENP-E motor on microtubules, did not abolish the bi-modal Gaussian distribution completely ()). These results indicate that full-length CENP-E with a long and flexible coiled-coil can sustain a two-conformational angular state relative to CENP-A at bi-oriented kinetochores, which is likely due to sister kinetochores associating with growing and shrinking ends of microtubules.
Figure 3. CENP-E displays a two-state conformation on bi-oriented kinetochores depending on microtubule dynamics and CENP-E motor activity. (a) Representative plots of CENP-E axis and CENP-A axis in a 3-dimensinoal space used to calculate the angular distribution of CENP-E along the kinetochore (CENP-A) axis. The red lines are representative lines depicting how the angular distribution was measured based on the CENP-E’s motor-to-tail axis and CENP-A-to-CENP-A axis in a sister kinetochore pair. Axes are shown in pixel (pixel = 67.1875 nm). (b–d) Angular distribution of FL-CENP-E in T98G cells treated with (b) DMSO, (c) Taxol and (d) GSK from three independent experiments (DMSO: n = 66, Taxol: n = 30, and GSK: n = 72). Plotted lines represent the robust non-linear regression fitting assuming the sum of two Gaussian distribution. The corresponding p-values from a Hartigans’ dip test analysis for multi-modality are shown (p values below 0.05 indicate the distribution is, at least, bimodal).
Chromosome misalignment caused by chemical inhibition of CENP-E can be rescued by inhibition of Aurora B kinase
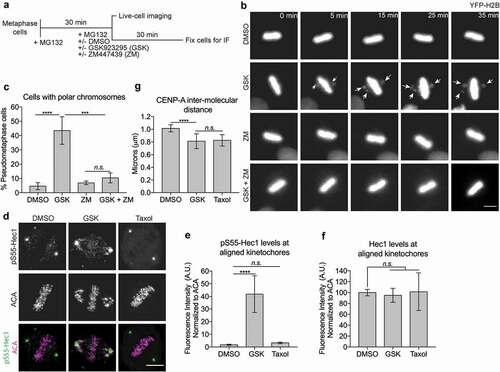
Full-length-CENP-E showed different conformations on unaligned and aligned kinetochores ()), suggesting possible different functional roles of CENP-E in response to initial kinetochore microtubule capture and bi-orientation of sister-kinetochores, respectively. It has been shown that CENP-E powers unaligned chromosomes to the metaphase plate [Citation5]. To better understand the functional roles of CENP-E on aligned kinetochores, we examined chromosome movement upon inhibition of CENP-E motor activity by GSK923295 in metaphase cells ()). Using live-cell imaging ()) and indirect immunofluorescence analysis of fixed cells ()), we found that polar chromosomes generated in metaphase cells by GSK923295 treatment were prevented by a simultaneous treatment of ZM447439, an Aurora B kinase inhibitor. Addition of GSK923295, but not ZM447439, to metaphase cells with fully aligned chromosomes produced misaligned chromosomes near spindle poles. By contrast, metaphase alignment was not disrupted upon co-treatment with GSK923295 and ZM447439. These results indicate a possible connection between the motor CENP-E and a signalling pathway involving Aurora B kinase at aligned kinetochores.
Figure 4. Maintaining low levels of Aurora B-mediated phosphorylation on attached kinetochores depends on the motor motility of CENP-E. (a) Schematic representation of chemical inhibition treatments protocol. (b) Aurora B inhibition can rescue chromosome misalignment caused by inhibition of CENP-E motility. Representative still frames of live-cell imaging of HeLa cells stably expressing YFP-H2B treated with or without a CENP-E inhibitor, GSK923295 (GSK), and an Aurora B inhibitor, ZM447439 (ZM), as indicated. Arrows indicate misaligned chromosomes. Scale bar, 20 μm. (c) Quantification (mean ± SD of three independent experiments) of T98G metaphase cells with fully aligned chromosomes or pseudo-metaphase cells with polar chromosomes after drug treatment (DMSO: n = 465, GSK: n = 487, ZM: n = 328, and GSK + ZM: n = 332). One-way ANOVA followed by a Tukey’s multiple comparison test was used to compare the means (*** p < 0.001, ****p < 0.0001, and n.s., not significant). (d) Inhibition of CENP-E motility results in an increase of Aurora B-mediated Hec1 phosphorylation. Immunofluorescence images showing Hec1 phosphorylation (pS55-Hec1) and ACA (anti-centromere antigen as the kinetochore marker) at aligned kinetochores. Scale bar, 5 μm. (e) Inhibition of CENP-E motor activity, but not microtubule dynamics, results in an increase of Aurora B-mediated phosphorylation of Hec1. Quantification (mean ± SD of three independent experiments) of normalized integrated intensity of pS55-Hec1 signals against ACA signals (DSMO: n = 223, GSK: n = 244, and Taxol: n = 202) on aligned kinetochores at the metaphase plate. One-way ANOVA followed by a Tukey’s multiple comparison test was used to compare the means (****p < 0.0001 and n.s., not significant). (f) Inhibition of CENP-E motor activity or microtubule dynamics does not affect kinetochore recruitment of Hec1. Quantification (mean ± SD of three independent experiments) of normalized integrated intensity of Hec1 signals against ACA signals on aligned kinetochores at the metaphase plate. More than 200 aligned kinetochores per group were quantified. An unpaired t test was used to compare the means (n.s., not significant). (g) A slight reduction of inter-kinetochore stretch (tension) caused by inhibition of CENP-E motor activity can be achieved by inhibition of microtubule dynamics. Quantification (mean ± SD of three independent experiments) of the distance between sister-kinetochore pairs (inter-kinetochore stretch) (DSMO: n = 215, GSK: n = 240, and Taxol: n = 214). One-way ANOVA followed by a Tukey’s multiple comparison test was used to compare the means (****p < 0.0001 and n.s., not significant).
To test the possible connection between CENP-E function and Aurora B-mediated phosphorylation at the kinetochore, we examined the levels of phosphorylated Hec1 (pS55-Hec1), a component of the Ndc80 complex and a major Aurora B substrate at outer kinetochores [Citation25]. This revealed higher levels of the kinetochore-associated pS55-Hec1 signals on chromosomes aligned at the metaphase plate upon addition of the CENP-E inhibitor, GSK923295 ()). By contrast, the kinetochore localization of Hec1 itself was not affected by this condition ()). To test whether the increase of pS55-Hec1 signal was due to the slight loss of tension (inter-kinetochore stretch) observed upon treatment with GSK923295, we treated cells with taxol, which affects both inter- and intra-kinetochore stretches [Citation21,Citation26]. Short-term treatment with taxol (5 min at 1 μM) resulted in a similar decrease in tension as treatment of GSK92395 ()), but did not cause elevated levels of pS55-Hec1 ()). Collectively, these results suggest a signalling pathway depending on the motor activity of CENP-E at aligned kinetochores to sustain low levels of Aurora B-mediated phosphorylation on outer kinetochore microtubule binding components, including the Ndc80 complex, maintaining kinetochore association with dynamic microtubule plus ends.
Low levels of Aurora B-mediated phosphorylation at bi-oriented kinetochores depends on coiled-coil-mediated conformational changes of CENP-E
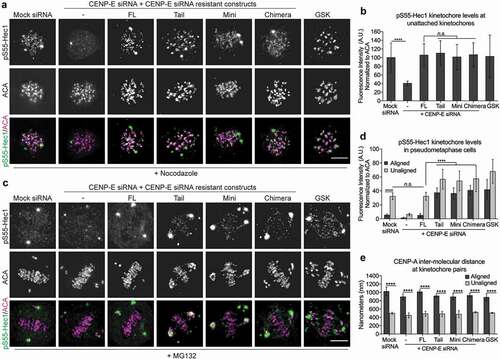
Phosphorylation of BubR1, a CENP-E binding partner at the kinetochore [Citation27], at Threonine 608 depends on CENP-E and is sensitive to kinetochore-microtubule attachment [Citation28]. Replacing endogenous BubR1 with a mutant that abrogates phosphorylation at Threonine 608 or depletion of CENP-E reduces the levels of Aurora B-mediated phosphorylation of Hec1 (pS55-Hec1) at unattached kinetochores [Citation28]. We found that cells expressing Mini- or Chimera-CENP-E, as well as Tail-CENP-E, rescued the levels of pS55-Hec1 as effective as the Full-length-CENP-E at unattached kinetochores ()). As inhibition of CENP-E by GSK923295 elevates Aurora B-mediated phosphorylation of the Ncd80 complex on attached kinetochores at the metaphase plate, we evaluated the levels of pS55-Hec1 in pseudo-metaphase with an obvious metaphase plate and a few polar chromosomes. This revealed that in contrast to the Full-length-CENP-E, both Mini- and Chimera-CENP-E behaved like Tail-CENP-E with increased levels of pS55-Hec1 on attached kinetochores at the metaphase plate and on unaligned kinetochores ()). Furthermore, in contrast to unattached kinetochores, the inter-kinetochore stretch (tension) on attached kinetochores was only reduced slightly in cells expressing CENP-E mutants in comparison to cells expressing Full-length-CENP-E ()). This slight decrease of tension was not significantly different from cells treated with GSK923295. Thus, there is a tension-independent mechanism to maintain low levels of Aurora B-mediated phosphorylation of Hec1 on aligned kinetochores that requires CENP-E function.
Figure 5. Maintaining low levels of Aurora B-mediated phosphorylation of outer kinetochore components on attached kinetochores depends on flexible conformational changes of CENP-E. (a) CENP-E tail domain is essential for Aurora B-mediated phosphorylation of Hec1 on unattached kinetochores. Immunofluorescence analysis of pS55-Hec1 and ACA in nocodazole treated T98G cells depleted of endogenous CENP-E, or expressing FL-CENP-E or CENP-E mutants as indicated. Scale bar, 5 μm. (b) Quantification (mean ± SD of three independent experiments) of the normalized integrated intensities of the kinetochore signals of pS55-Hec1/ACA (Mock siRNA: n = 215, CENP-E siRNA: n = 222, FL: n = 219, Tail: n = 203, Mini: n = 226, Chimera: n = 230, and GSK n = 212). An unpaired t test or a One-way ANOVA followed by a Tukey’s multiple comparison test was used to compare the means (****p < 0.0001 and n.s., not significant). (c) Cells expressing Full-length CENP-E, but not Tail-, Mini-, or Chimera-CENP-E, can maintain low levels of Hec1 phosphorylation on bi-oriented kinetochores. Immunofluorescence analysis of pS55-Hec1 and ACA in MG132-treated pseudo-metaphase T98G cells. Scale bar, 5 μm. (d) Quantification (mean ± SD of three independent experiments) of the normalized integrated intensities of the kinetochore signals of pS55-Hec1/ACA on aligned and unaligned kinetochores (aligned, Mock siRNA: n = 249, CENP-E siRNA: n = 233, FL: n = 193, Tail: n = 207, Mini: n = 197, Chimera: n = 171, and GSK n = 215; unaligned, Mock siRNA: n = 119, CENP-E siRNA: n = 199, FL: n = 100, Tail: n = 134, Mini: n = 118, Chimera: n = 191, and GSK n = 223). An unpaired t test or a One-way ANOVA followed by a Tukey’s multiple comparison test was used to compare the means (****p < 0.0001 and n.s., not significant). (e) CENP-E is not required for inter-kinetochore stretch (tension) upon bio-orientation. Quantification (mean ± SD of three independent experiments) of distance between sister kinetochore pairs (inter-CENP-A stretch) of aligned and unaligned kinetochores. More than 30 kinetochores pairs per group were quantified. An unpaired t test or a One-way ANOVA followed by a Tukey’s multiple comparison test was used to compare the means (****p < 0.0001).
A tension-independent mechanism to reduce Aurora B-mediated phosphorylation at the kinetochore in response to microtubule capture by CENP-E
To test the existence of a tension-independent mechanism to maintain low levels of Hec1 phosphorylation by Aurora B kinase, we examined the levels of kinetochore-associated pS55-Hec1 in mono-polar spindles upon treatment with monastrol, an Eg5 inhibitor that prevents the separation of spindle poles [Citation17]. The mono-polar spindle generates monotelic attachments, in which one sister kinetochore captures microtubules from spindle poles, whereas the other sister kinetochore remains unattached ()). Analysis of Aurora B phosphorylation using an established FRET probe [Citation24] showed an asymmetric levels of phosphorylation on monotelic sister kinetochores in mono-polar spindles, which was sensitive to the treatment of ZM447439, the inhibitor of Aurora B kinase (Figure S8, a and b). The asymmetric attachment status of these monotelic kinetochores was confirmed using indirect immunofluorescence of Mad1 (Figure S8, c and d). The distal kinetochore showed an increased Mad1 signal relative to the proximal in pairs of sister kinetochores, suggesting that the distal kinetochore is unattached and the proximal kinetochore is attached to microtubules in a pair of sister kinetochores. This asymmetric pattern of monotelic sister kinetochores was similar when we immunostained for CENP-E (Figure S8, c and e). Replacing endogenous CENP-E with Mini-, Chimera-, or Tail-CENP-E did not altered the asymmetric localization pattern of CENP-E on monotelic sister kinetochores (Figure S8f). These results collectively demonstrate the existence of a tension-independent microtubule attachment pathway at the kinetochore that regulates Aurora B-mediated phosphorylation of outer kinetochore components.
Figure 6. A tension-independent mechanism to reduce Aurora B phosphorylation at the kinetochore upon microtubule capture by CENP-E. (a) Diagram depicting the three types of microtubule-kinetochore attachments in monastrol-treated cells. (b) CENP-E levels are asymmetric in monotelic-attached sister-kinetochore pairs. Immunofluorescence images of monastrol-treated cells were stained with antibodies against CENP-E, ACA, and tubulin (microtubules). Scale bar, 5 μm. Insets represent a pair of monotelic-attached sister kinetochores. (c) The levels of pS55-Hec1 are asymmetric in monotelic-attached sister-kinetochore pairs, which depends on normal CENP-E function. Monastrol-treated cells were stained with antibodies against ACA, CENP-E, and pS55-Hec1. Scale bar, 5 μm. Insets represent a pair of monotelic-attached sister kinetochores. (d) Quantification (mean ± SD of three independent experiments) of normalized integrated fluorescence intensity of pS55-Hec1/ACA signals on monotelic kinetochores (Mock siRNA: n = 49, CENP-E siRNA: n = 50, FL: n = 39, Tail: n = 32, Mini: n = 42, and Chimera: n = 35). An unpaired t test was used to compare the means (****p < 0.0001 and n.s., not significant). (e) Quantification (mean ± SD of three independent experiments) of normalized integrated fluorescence intensity of Hec1/ACA signals on monotelic kinetochores (Mock siRNA: n = 35, CENP-E siRNA: n = 31, FL: n = 32, Tail: n = 31, Mini: n = 32, and Chimera: n = 31). An unpaired t test was used to compare the means (n.s., not significant).
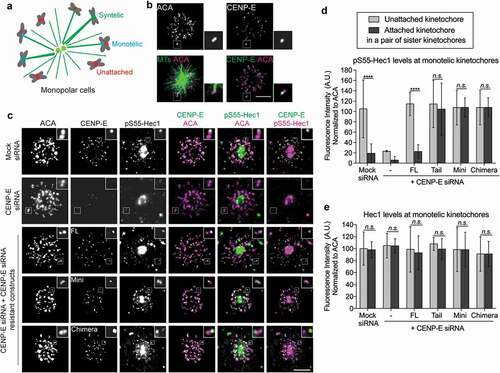
To further examine a possible role of CENP-E involved in a tension-independent pathway regulating Aurora B-mediated phosphorylation at the kinetochore, we assessed the effects of CENP-E mutants on the levels of pS55-Hec1 at monotelic sister kinetochores in mono-polar spindles. The kinetochore-associated pS55-Hec1 levels exhibited similar asymmetry as the Aurora B phosphorylation FRET probe on monotelic sister kinetochores ()). By contrast, the total Aurora B levels (Figure S9) and Hec1 levels regardless of its phosphorylation status ()) remained the same on both sister kinetochores. Replacing endogenous CENP-E with Mini-, Chimera-, or Tail-CENP-E altered asymmetric levels of pS55-Hec1, in which Hec1 remained highly phosphorylated on attached sister kinetochores even with decreased levels of CENP-E ()). Taken together, these results demonstrate the existence of a tension-independent mechanism to reduce Aurora B-mediated phosphorylation of the Ndc80 complex in response to microtubule capture by CENP-E and, thus, to facilitate the stabilization of kinetochore-microtubule attachments before tension generation.
Discussion
Current models for regulation of Aurora B-mediated phosphorylation of outer kinetochore components emphasize the distance (tension or inter-kinetochore stretch) between sister-kinetochores to move inner centromere localized Aurora B away from its substrates (e.g. the Ndc80 complex) at outer kinetochores [Citation1,Citation29]. The asymmetric Hec1 phosphorylation levels exhibited on monotelic sister kinetochores () clearly suggests the existence of other mechanisms independent of tension regulating Aurora B-mediated phosphorylation of outer kinetochore components. It has been shown that active Aurora B is able to discriminate between correct and incorrect attachments regardless of its localization clustering on either centromeric chromatin or microtubules [Citation30]. Besides an inter-kinetochore stretch (tension), an intra-kinetochore stretch has also been observed upon microtubule attachment [Citation26,Citation31], which could represent structural changes within the kinetochore [Citation20]. However, the intra-kinetochore stretch is not necessary to mediate stabilization of kinetochore-microtubule attachments [Citation21,Citation32,Citation33]. Our findings support the existence of a tension-independent mechanism reducing Aurora B-mediated phosphorylation in response to microtubule capture by the kinetochore-associated plus-end motor CENP-E in human cells.
Several functional roles, which are not mutually exclusive, have been demonstrated for the kinetochore-associated kinesin motor CENP-E: (1) CENP-E is an essential motile tether between kinetochores and microtubules [Citation9,Citation10,Citation13,Citation14]; (2) CENP-E transports misaligned (polar) chromosomes to the metaphase plate [Citation5]; (3) CENP-E is a processive bi-directional tracker of dynamic microtubule ends [Citation6]; and (4) CENP-E regulates the mitotic checkpoint through its interaction with BubR1 [Citation27,Citation28,Citation34–Citation37]. Our results demonstrate another functional role of kinetochore-associated CENP-E to maintain low levels of Aurora B-mediated phosphorylation of outer kinetochore microtubule binding proteins (e.g. Hec1) and, thus, to enhance the link between kinetochores and the dynamic microtubule plus ends. Inhibition of CENP-E motor activity produces elevated Aurora B phosphorylation and misaligned chromosomes, which can be rescued by inhibiting Aurora kinase activity (). This may involve the recruitment of phosphatases to regulate the balance of phosphorylation/dephosphorylation. Indeed, a previous study [Citation12] has shown that Aurora B-mediated phosphorylation of CENP-E regulates microtubule capture by CENP-E through a phosphatase targeting pathway involving the Protein Phosphatase 1, PP1. More importantly, the kinetochore PP1 targeting motif is found at the coiled-coil domain near the motor domain of CENP-E. Furthermore, BubR1, the CENP-E binding partner, has also been shown to associate with B56-PP2A phosphatase at the kinetochore [Citation38]. Phosphorylation of BubR1 itself as well as its effects of Aurora B-mediated phosphorylation depend on CENP-E and its microtubule binding status [Citation28]. An alternative explanation to the kinases/phosphatases balance would be that inhibition of CENP-E motor activity on bi-oriented kinetochores results in less stable kinetochore-microtubule attachment and, therefore, an increase of Aurora B-mediated phosphorylation. However, this is less likely as tension (the inter-kinetochore stretch) between sister-kinetochores remains high () and )) and Mad1 remains low at aligned kinetochores (Figure S4) in cells replacing endogenous CENP-E with Mini- or Chimera-CENP-E or chemical inhibition of CENP-E motor activity.
Previous studies on the configuration of kinetochore components identified a bent and rigid lateral linkage of the Ndc80 complex through an elongated interaction with the Mis12 complex and KNL1 [Citation20]. This study proposed the existence of a flexible linkage that could transmit the pulling forces generated by curling protofilaments of microtubule to the inner kinetochore at bi-oriented kinetochores. No such linkage in vertebrate cells has so far been identified. Analysis of the angular distribution of CENP-E along the kinetochore yielded a bi-modal distribution on a pair of bi-oriented sister kinetochores (). GSK-923295, a specific allosteric inhibitor of CENP-E, locks CENP-E with attached microtubules in the original bi-modal for short-term treatment. In vitro studies have characterized CENP-E as a microtubule tip-tracker, tracking both the growing and shrinking end of microtubules [Citation6]. Thus, we propose that CENP-E undergoes a two-state conformational change acting as a flexible linkage that helps track the plus ends of both growing and shrinking microtubules. This mechanism depends on the motor activity and the elongated, flexible coiled-coil of CENP-E. Neither Mini- nor Chimera-CENP-E exhibits the bi-modal distribution at the aligned kinetochores. The much shorter Mini-CENP-E at the kinetochore may have difficulty to track shrinking microtubule plus ends, whereas the Chimera-CENP-E with an extended conformation cannot track the plus tips of growing microtubules.
Materials and methods
Tissue culture, transfection and drug treatment
T98G and HeLa cells were cultured in DMEM supplemented with 10% Fetal Bovine Serum at 37°C in 5% CO2. Co-transfections of the CENP-E siRNA (5ʹ - AGAUAAGGGAACAGGAAAUUU - 3ʹ) and GFP-tagged, double-tagged mCherry and EmeraldGFP CENP-E constructs or Aurora B phosphorylation FRET biosensors transgenes into T98G cells were performed using DharmaFECT Duo Transfection Reagent (GE Dharmacon) according to the manufacturer’s instruction. 48 hrs after transfection, cells were synchronized with 100 μM monastrol (Sigma-Aldrich) for 4 hrs then either fixed or released into 10 μM MG132 for 20–30 min. Nocodazole (Sigma-Aldrich) treatment was performed at 3.3 μM for 4 hrs. For live-cell imaging experiments, HeLa cells stably expressing YFP-H2B were treated 10 μM MG132 (Sigma-Aldrich) for 30 min prior to imaging then treated with 10 μM MG132 with or without 50 nM GSK923295 (Selleckchem) and/or 2 μM ZM447434 (Tocris Bioscience) upon imaging or fixed after 30 min. Taxol (Tocris Bioscience) treatment was performed at 1 μM for 5 min.
Plasmid construction
Human Full-length-CENP-E sequence (1-2698aa), Tail-CENP-E (1572aa-2698aa), Mini-CENP-E (Motor domain 1-470aa plus the tail domain 1572aa-2698aa) were amplified using PCR and subsequently sub-cloned into pEGFP-C1 plasmid (Clontech Laboratories, Inc.) between NheI and SalI cloning restriction sites. To construct Chimera-CENP-E, the coiled-coil domain of human Kinesin 5B (318aa – 913aa) was amplified using PCR and then sub-cloned into the XhoI site of the Mini-CENP-E-GFP construct. To generate double tagged-CENP-E constructs, the Gateway system (Invitrogen) was used. FL-CENP-E, Mini-CENP-E and Chimera-CENP-E sequences were amplified using PCR with the TOPO-cloning reaction (Invitrogen) and introduced into the pENTR/D-TOPO entry cloning vector (Invitrogen), then subsequently sub-cloned into an mCherry-GW-EmeraldGFP vector (kindly gifted by Geri Kreitzer from The City University of New York) using a LR Clonase II reaction (Invitrogen). All constructs were verified by sequencing and DNA was purified with maxiprep kits (QIAGEN).
Antibody labelling
Phospho-Hec1 (pS55-Hec1) and ACA antibodies were purchased from GeneTex and Antibodies Incorporated, respectively. CENP-E and Mad1 antibodies were from Santa Cruz. All other antibodies were purchased from Abcam. Cells were grown on poly-l-lysine–coated No. 1.5 coverslips and fixed with −20°C Methanol for 10 min. Prior to phospho-staining of pS55-Hec1, cells were pre-extracted with 0.1% Triton X-100 (Fisher Scientific) in microtubule stabilizing buffer, MTSB (100 mM PIPES, 1 mM EGTA, 1 mM MgSO4, and 30% of glycerol) for 30–90 sec and immediately fixed with −20°C Methanol for 10 min. Fixed cells were blocked in PBS containing 0.1% Triton X-100 and 5% Bovine Serum Albumin (Sigma-Aldrich) for 1 hr at room temperature or overnight at 4°C. Coverslips were subjected to primary antibodies diluted in blocking buffer for 1 hr at room temperature or overnight at 4°C, and then to secondary antibodies conjugated to Alexa 488 (Invitrogen), Rhodamine or Cy5 (Jackson Immuno-Research Laboratories, Inc.) for 45 min at room temperature, and finally followed by DAPI counter-staining. Coverslips were mounted with antifade reagent (SouthernBiotech).
Image acquisitions
Image acquisitions were performed at room temperature using an inverted microscope (IX81; Olympus) with a 60X, NA 1.42 Plan Apochromat oil immersion objective lens (Olympus) and a monochrome charge-coupled device camera (Sensicam QE; Cooke Corporation). The SlideBook imaging software (Intelligent Imaging Innovations, 3i) was used to acquire the 500 nm Z-sections or 200 nm Z-sections for the K-SHREC analysis of CENP-E. To quantify fluorescence intensities, all images were collected on the same day using identical exposure times. For live-cell imaging experiments, HeLa cells were plated onto 4 compartment glass-bottom dishes No. 1.5 (Greiner Bio-One) and imaged at 37°C in 5% CO2. Images were acquired using a 40X, NA 0.6 dry objective lens using the SlideBook imaging software. Cells expressing the Aurora B phosphorylation biosensor were imaged lived using a Nikon Ti Eclipse inverted microscope with a 100x NA 1.49 oil immersion objective lens and the NIS-Elements (Nikon) software.
Data and statistical analysis
Quantitative analysis of raw 16-bit tiff stacks or maximum Z-projected fluorescence images was performed using ImageJ (NIH). Kinetochore fluorescence intensities were quantified by drawing a small 6 × 6 pixel and a large 12 × 12 pixel circular regions centered over each kinetochore to obtain the total integrated fluorescence of each region. The final total integrated fluorescence was obtained by subtracting the total integrated fluorescence of the large circular region from the smaller total integrated fluorescence. This local background subtraction method controls for background fluorescence heterogeneity. The fluorescence intensities were normalized to the total integrated fluorescence of ACA and then normalized to the total integrated fluorescence obtain from nocodazole-treated cells.
The centroid positions of kinetochores and centrosomes were obtained by drawing a circular mask over the ACA and γ-tubulin fluorescence signals, respectively. The X, Y and Z coordinates of mCherry-CENP-E-EmGFP constructs and CENP-A centroids were determined by a nonlinear curve fitting function of segmented kinetochore volume with 3D Gaussian fitting in MATLAB (lsqcurvefit, R2016a; MathWorks). The X, Y and Z coordinates were then corrected for chromatic aberration using 100 nm multi-coated fluorescent microspheres (Fisher Scientific).
The inter-kinetochore stretch was measured as previously described [Citation22] by using line-scans of CENP-A immunofluorescence signal of sister kinetochore pairs in the same focal plane to obtain the distance between the brightest pixels of the kinetochore pairs.
Representative images were subjected to no-neighbour deconvolution using Slidebook imaging software and maximum Z-projection of selected stacks and subsequently scaled equally in all conditions tested in ImageJ (NIH), as we described previously [Citation39].
Statistical analyses were performed using GraphPad Prism (version 7a) using unpaired, two-tailed t tests to compare the means between two groups or a One-way ANOVA was used to compare the means of 3 or more groups as specified in the figure legends. Hartigans’ dip test statistical analysis (version 0.75–7) was performed in R (version 1.1.463). Akaike Information Criterion (AIC) analysis was conducted in Matlab to assess which Gaussian Mixture Model best fits (in terms of the number of components) per each sample. Plots were prepared in GraphPad Prism (version 7.0a) or in Matlab (version R2016a).
Supplemental Material
Download Microsoft Video (AVI) (1 MB)Supplemental Material
Download Microsoft Video (AVI) (1 MB)Supplemental Material
Download Microsoft Video (AVI) (1 MB)Supplemental Material
Download Microsoft Video (AVI) (1 MB)Supplemental Material
Download MP4 Video (667.2 KB)Supplemental Material
Download Zip (14.2 MB)Acknowledgments
We thank all members of the Mao laboratory, especially Yige Guo for technical support and stimulating discussions. We thank Jennifer Deluca, Michael Lampson and Geri Kreitzer for reagents. We thank Theresa Swayne and Adam White at Columbia University Herbert Irving Comprehensive Cancer Center Shared Resources for technical support on FRET analysis. We are grateful to the quantitative training that C.T. received from the Quantitative Imaging course (2015) at Cold Spring Harbor Laboratory and that C.L. received from the Physiology Course (2013) at Marine Biological Laboratory. This work has been supported by a grant from the National Institute of Health (GM89768) to Y.M. C.T. has been supported by a Research Supplement to Promote Diversity in Health-Related Research from NIH/NIGMS and a T32 Cancer Biology Training grant by Columbia University Herbert Irving Comprehensive Cancer Center.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140.
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158.
- Yoo TY, Choi JM, Conway W, et al. Measuring NDC80 binding reveals the molecular basis of tension-dependent kinetochore-microtubule attachments. Elife. 2018;7:e36392.
- Saurin AT. Kinase and phosphatase cross-talk at the kinetochore. Front Cell Dev Biol. 2018;6:62.
- Kapoor TM, Lampson MA, Hergert P, et al. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391.
- Gudimchuk N, Vitre B, Kim Y, et al. Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat Cell Biol. 2013;15:1079–1088.
- Lombillo VA, Nislow C, Yen TJ, et al. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995;128:107–115.
- Martin-Lluesma S, Stucke VM, Nigg EA. Role of hec1 in spindle checkpoint signaling and kinetochore recruitment of mad1/mad2. Science. 2002;297:2267–2270.
- McEwen BF, Chan GK, Zubrowski B, et al. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12:2776–2789.
- Putkey FR, Cramer T, Morphew MK, et al. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3:351–365.
- Yao X, Anderson KL, Cleveland DW. The microtubule-dependent motor centromere-associated protein E (CENP- E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J Cell Biol. 1997;139:435–447.
- Kim Y, Holland AJ, Lan W, et al. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142:444–455.
- Kim Y, Heuser JE, Waterman CM, et al. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419.
- Yao X, Abrieu A, Zheng Y, et al. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491.
- Wood KW, Lad L, Luo L, et al. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc Natl Acad Sci U S A. 2010;107:5839–5844.
- Vitre B, Gudimchuk N, Borda R, et al. Kinetochore-microtubule attachment throughout mitosis potentiated by the elongated stalk of the kinetochore kinesin CENP-E. Mol Biol Cell. 2014;25:2272–2281.
- Kapoor TM, Mayer TU, Coughlin ML, et al. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988.
- Barisic M, Aguiar P, Geley S, et al. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat Cell Biol. 2014;16:1249–1256.
- Churchman LS, Okten Z, Rock RS, et al. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc Natl Acad Sci U S A. 2005;102:1419–1423.
- Wan X, O’Quinn RP, Pierce HL, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684.
- Magidson V, He J, Ault JG, et al. Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells. J Cell Biol. 2016;212:307–319.
- Waters JC, Chen RH, Murray AW, et al. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191.
- Thrower DA, Jordan MA, Wilson L. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motility Cytoskeleton. 1996;35:121–133.
- Liu D, Vader G, Vromans MJ, et al. Sensing chromosome bi-orientation by spatial separation of Aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353.
- DeLuca JG, Gall WE, Ciferri C, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982.
- Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381.
- Chan GK, Jablonski SA, Sudakin V, et al. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J Cell Biol. 1999;146:941–954.
- Guo Y, Kim C, Ahmad S, et al. CENP-E-dependent BubR1 autophosphorylation enhances chromosome alignment and the mitotic checkpoint. J Cell Biol. 2012;198:205–217.
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37.
- Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 2013;497:118–121.
- Uchida KS, Takagaki K, Kumada K, et al. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–390.
- Etemad B, Kuijt TE, Kops GJ. Kinetochore-microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint. Nat Commun. 2015;6:8987.
- Tauchman EC, Boehm FJ, DeLuca JG. Stable kinetochore-microtubule attachment is sufficient to silence the spindle assembly checkpoint in human cells. Nat Commun. 2015;6:10036.
- Abrieu A, Kahana JA, Wood KW, et al. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 2000;102:817–826.
- Mao Y, Abrieu A, Cleveland DW. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98.
- Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J Cell Biol. 2005;170:873–880.
- Weaver BA, Bonday ZQ, Putkey FR, et al. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563.
- Kruse T, Zhang G, Larsen MS, et al. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J Cell Sci. 2013;126:1086–1092.
- Liu C, Zhu R, Mao Y. Nuclear actin polymerized by mDia2 confines centromere movement during CENP-A loading. iScience. 2018;9:314–327.
