ABSTRACT
Several lines of evidence suggest that circular RNAs (circRNAs) play important roles in oncogenesis and tumor progression. However, our knowledge of the role of circRNAs in gastric cancer (GC) remains limited. We investigated the possibility that circular RNA 0047905 (circRNA0047905) might act as a tumor promoter in the pathogenesis of gastric cancer by profiling miRNA expression in GC tissues and paired noncancerous mucosa tissues using miRNA microarrays. Next, a ceRNA network was constructed according to common miRNAs binding circRNAs and mRNAs. Mechanistically, we demonstrated that circRNA0047905 directly binds miR4516 and miR1227-5p, relieving suppression for targets SERPINB5 and MMP11. We observed that down-regulated circRNA0047905 expression in gastric cancer cells inhibited Akt/CREB signaling pathway activation. RNA scope in situ hybridization revealed expression of circRNA0047905 in GC. Our data suggest that circRNA0047905 is a promising target for GC diagnosis and therapy.
Introduction
Gastric cancer (GC) is the tumor fifth common malignant tumor, and is the third leading cause of death worldwide [Citation1]. According to high-quality data from the National Central Cancer Registry of China, an estimated 697,100 new gastric cancer cases and 498,000 gastric cancer deaths occurred in China in 2015 [Citation2]. Despite many advances in the diagnosis and treatment of this disease, the prognosis of patients with GC remains poor, with a 5-year overall survival of less than 30% in most countries [Citation3]. Cancer is widely regarded as a genetic disease, and GC is no exception. Nevertheless, the molecular mechanisms underlying the development and progression of GC, especially genetic and epigenetic changes, are poorly understood. Therefore, the discovery of new molecular mechanisms and therapeutic targets that may control the severity of GC and may provide predictive value for prognosis are of great importance.
Noncoding RNAs play important roles in cancer biology, providing potential targets for cancer intervention. As a new class of endogenous noncoding RNAs, circular RNAs (circRNAs) have been recently identified in cell development and function [Citation4]. Several years ago, Pandolfi et al. put forward the competing endogenous RNA (ceRNA) hypothesis that mRNA, lncRNA, pseudogene transcripts competitively bind to microRNA response elements (MREs) to regulate gene expression mutually, revealing a new mechanism of interaction among RNA transcripts [Citation5]. Some circRNAs and their corresponding linear RNAs are derived from the same exon, suggesting that circRNA might function as a new member of the ceRNA family [Citation6]. Importantly, a growing number of studies have demonstrated that circRNAs are closely associated with disease and may play significant roles in the pathogenesis and diagnosis of disease [Citation7–Citation9].
Identifying the main abnormally expressed by circRNAs and their roles in cancer has attracted much attention. Nevertheless, our knowledge of circRNAs in gastric cancer (GC) remains limited. In previous studies, we analyzed the expression profile of human circRNAs and mRNAs in GC tissues and identified that circRNA0047905 may act as a tumor promoter in the pathogenesis of GC [Citation10]. In this study, in order to elucidate the molecular mechanism of this circRNA in GC, we analyzed the differential expression profiles of human miRNAs between GC tissue and paired noncancerous mucosa tissues with miRNA microarrays. Through bioinformatics analysis and a luciferase assay system, we found that circRNA0047905 was a ceRNA for miR4516 and miR1227-5p, initiating gastric cancer progression. Our findings reveal a novel mechanism underlying circRNA0047905 in GC progression.
Results
MicroRNA expression profiles in GC
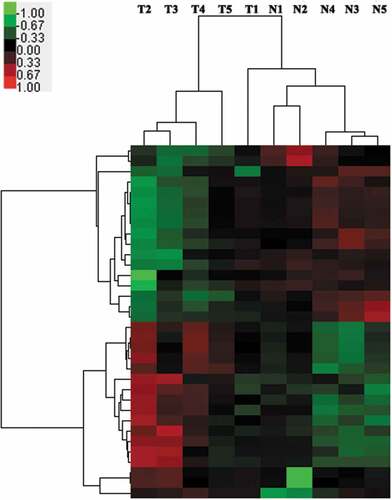
High-throughput microarrays is an efficient approach for investigating the biological function of RNAs. The Agilent Technology Human miRNA Microarray (V 21.0, 8 × 60 K) was used to profile miRNA expression, in five paired gastric cancer tissues and adjacent normal tissues (). In total, 2,549 miRNAs transcripts were collected from authoritative databases such as miRbase. To study these microarray data and to discover potential biomarkers not yet mined completely, stringent filtering criteria were used (raw data ≥ 10, fold-change ≥ 2, FDR < 0.05) [Citation11]. We identified thirty-four miRNAs that were differentially-expressed in gastric cancer tissues versus adjacent normal tissues. Seventeen of these miRNAs were upregulated, and the remaining seventeen miRNAs were downregulated (Supplemental Table S1).
Figure 1. Heat map of the miRNA profiles in GC tissues versus paired normal gastric mucosa tissues.
Prediction of circRNAs related to GC and target genes
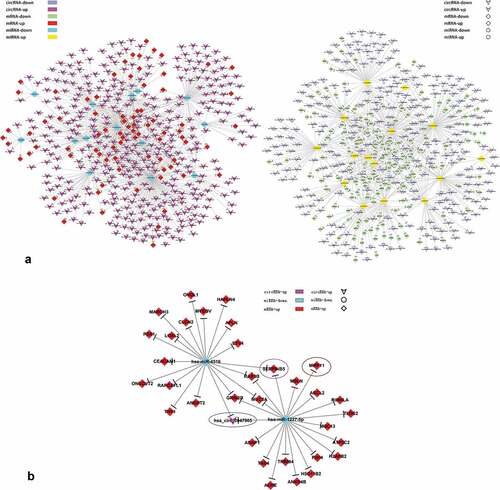
Based on the results of aberrantly-expressed RNAs obtained from the previous experimental data, TargetScan and miRanda databases were used to investigate the potential targets of miRNAs, and the ceRNA network was constructed according to the common miRNAs binding circRNAs and mRNAs [Citation12]. By merging the common targeted miRNAs, we constructed a network of circRNAs-miRNAs-mRNAs with a total of 591 circRNAs, 223 mRNAs and 29 miRNAs (T vs N; Fold-change ≥ 2 or Fold-change ≤ 0.5, FDR< 0.05) (). Each gene corresponded to a node, and two genes were connected by a string. This indicated the tight correlation and regulation relationship of these genes. In the network, we discovered that circRNA0047905 was a ceRNA for miR4516 and miR1227-5p, promoting tumorigenesis by stimulating expression of SERPINB5 and MMP11 ().
Figure 2. circRNAs-miRNAs-mRNAs network.
Overexpression of SERPINB5 and MMP11 correlated with poor prognosis in GC patients
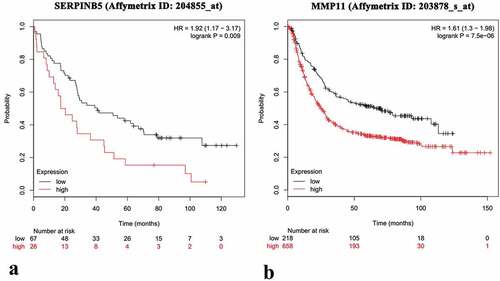
This experiment highlighted the importance of SERPINB5 and MMP11, because gastric tumor specimens showed increased SERPINB5 and MMP11 expression levels compared with those of corresponding normal tissues. In addition, the frequency of induction of both SERPINB5 and MMP11 was associated with the stage of gastric cancer (GC) and lymph node metastasis [Citation13–Citation16]. Via the Kaplan-Meier Plotters database, we found that the frequency of SERPINB5 and MMP11 induction was associated with poor prognosis in GC patients [Citation17] ().
Figure 3. The Kaplan-Meier survival analysis of SERPINB5 and MMP11 in GC patients.
Correlation between circRNA0047905 and SERPINB5 and MMP11 in GC
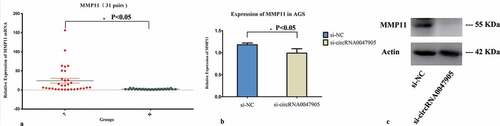
In a previous study, we observed both circRNA0047905 and SERPINB5 mRNA were up-regulated in GC tissues and our data supported the notion that there was a significantly positive correlation between circRNA0047905 and SERPINB5 mRNA in GC tissues. Meanwhile, mRNA expression levels of SERPINB5 were downregulated in AGS GC cells when using siRNA to interfere with the expression of circRNA0047905 [Citation10]. In this study, we examined the expression MMP11 by qRT-PCR in 31 matched GC and adjacent non-cancer tissues. Gastric tumor specimens showed greater MMP11 expression levels than those of corresponding normal tissues (). We examined MMP11 mRNAs expression levels for specific siRNA-targeted circRNA0047905 in AGS cells and compared them with the negative control group. The mRNA expression levels of MMP11 () were downregulated in the si-circRNA0047905 cell group, and protein expression level of MMP11 was significantly downregulated in the si-circRNA0047905 group (). These data suggested that circRNA0047905 may regulate SERPINB5 and MMP11 expression in gastric cancer cells.
Figure 4. Relative expression of MMP11 in human GC cell lines transfected with specific siRNA targeted circRNA0047905.
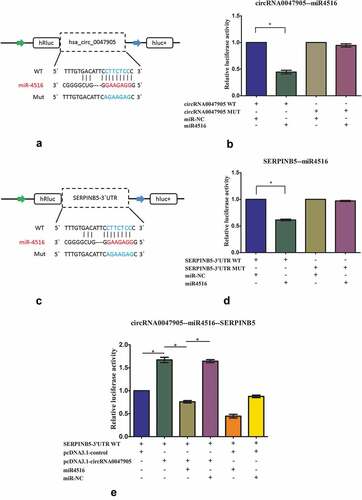
Effects of circRNA0047905 on SERPINB5 expression as a ceRNA
According to miRBase prediction, circRNA0047905 possessed a complementary sequence to miR4516 seed regions. MiRNAs suppress translation and degrade mRNA in an AGO2-dependent manner by binding to their targets. Luciferase reporters were constructed by inserting either the wild-type circRNA0047905 sequence (WT) or the sequence with mutated binding sites of miR4516 (Mut) into the 3ʹUTR of renilla luciferase (). Upregulation of miR4516 remarkably reduced luciferase activity of the WT reporter but not of the mutant reporter (), suggesting that miR4516 interacted with circRNA0047905 via a complementary seed region. According to miRBase prediction, miR4516 targeted SERPINB5 3ʹUTR with a high score. Next, luciferase reporter assays were performed to verify this interaction. Transfection of miR4516 mimics substantially reduced the activity of a luciferase reporter carrying the SERPINB5-3ʹUTR of WT compared to that of miR-NC (). These results suggested that miR4516 targeted SERPINB5 and negatively regulated its expression. In order to further explore the molecular mechanisms underlying circRNA0047905, we co-transfected the luciferase reporter of WT SERPINB5 3ʹ UTR with pcDNA3.1-circRNA0047905 overexpression vector. Luciferase activity of WT SERPINB5 3ʹ UTR with pcDNA3.1-circRNA0047905 overexpression vector was significantly greater than that of the control group of co-transfection with WT luciferase reporter and pcDNA3.1-control, suggesting that overexpression of circRNA0047905 increased luciferase activity of WT SERPINB5 3ʹUTR reporter by binding endogenous miR4516. This effect could be evidently abolished by overexpression of miR4516 (). These data suggested that circRNA0047905 as a ceRNA for miR4516 stimulated the expression of SERPINB5.
Figure 5. Overexpression of circRNA0047905 increased luciferase activity of SERPINB5-3ʹUTR-WT reporter by binding endogenous miR4516.
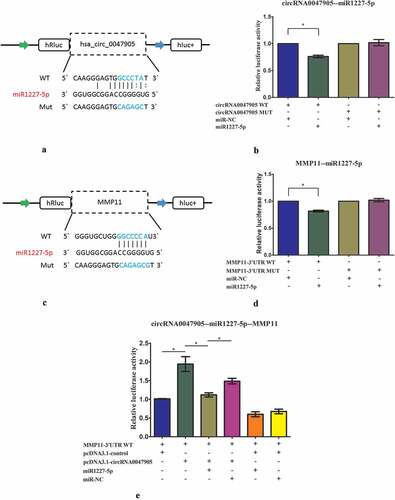
Effects of circRNA0047905 on MMP 11 expression as a ceRNA
According to miRBase prediction, circRNA0047905 possessed a complementary sequence to miR1227-5p seed regions. As circRNAs could function as miRNA sponges that naturally sequester and competitively suppress miRNA activity, we predicted that circRNA0047905 functioned as a decoy to regulate MMP11 expression through a similar mechanism. Luciferase reporters were constructed by inserting either the wild-type circRNA0047905 sequence (WT) or the sequence with mutated binding sites of miR1227-5p (Mut) into the 3ʹUTR of renilla luciferase (). Upregulation of miR1227-5p reduced luciferase activities of the WT reporter but not of the mutant reporter (), suggesting that miR1227-5p interacted with circRNA0047905 via a complementary seed region. According to miRBase prediction, miR1227-5p targeted MMP11 3ʹUTR with a high score. Next, luciferase reporter assays were performed to verify this interaction. Transfection of miR1227-5p mimics substantially reduced the activity of a luciferase reporter carrying the MMP11-3ʹUTR of WT compared to miR-NC (). These results indicated that miR1227-5p targeted MMP11 and negatively regulated its expression. In order to further explore the molecular mechanisms underlying circRNA0047905, we co-transfected the luciferase reporter of WT MMP11 3ʹ UTR with pcDNA3.1-circRNA0047905 overexpression vector. Luciferase activity of WT MMP11 3ʹ UTR with pcDNA3.1-circRNA0047905 overexpression vector was significantly greater than that of the control group of co-transfection with WT luciferase reporter and pcDNA3.1-control, suggesting that overexpression of circRNA0047905 increased luciferase activity of WT MMP11 3ʹUTR reporter by binding endogenous miR1227-5p. This effect was substantially abolished by overexpression of miR1227-5p (). These data suggested that circRNA0047905 as a ceRNA for miR1227-5p stimulated the expression of MMP11.
Figure 6. Overexpression of circRNA0047905 increased luciferase activity of MMP11-3ʹUTR-WT reporter by binding endogenous miR1227-5p.
Down-regulated circRNA0047905 expression in gastric cancer cells inhibits Akt/CREB signaling pathway activation
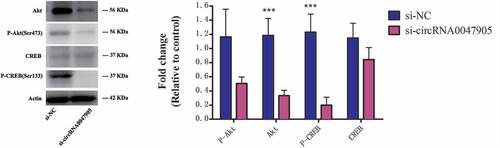
To comprehensively obtain mechanistic insights into the role of circRNA0047905 in regulating gastric cancer cell proliferation, we generated a phosphoprotein antibody array to identify ectopically-phosphorylated proteins induced in AGS gastric cancer cells transfected with siRNAs targeting circRNA0047905 and compared them with control. We identified a spectrum of proteins whose phosphorylation levels were altered by more than 20% when circRNA0047905 was down-regulated, including Akt, CREB, BAD, Hsp90, P53, and others (Supplemental Figure S1). Many of these proteins, when ectopically phosphorylated, are important for cell proliferation, migration, invasion and apoptosis. Among these proteins, we confirmed by western blot that targeted down-regulation of circRNA0047905 resulted in reduced phosphorylation of CREB and decreased expression levels of Akt ().
RNA scope in situ hybridization reveals the expression of circRNA0047905 in GC
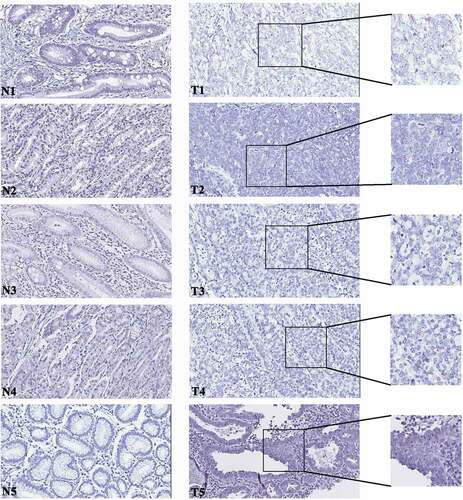
CircRNAs in tissue samples have been demonstrated to associate with tumor invasion, metastasis and patient prognosis. To assess the circRNA0047905 expression pattern in GC specimens, an RNA scope assay was performed in five GC tissue samples and paired adjacent noncancerous mucosal tissue samples [Citation18,Citation19]. BaseScopeTM probe is a 1ZZ probe targeting circRNA0047905 back-spliced junction (has_circ_0047905, 2173–37). This circRNA sequence matches human SERPINB5 exon 4 to exon 7 (NM_002639.4, 449–2633); therefore, the probe target region is equivalent to NM_002639.4 2621–485. The signal was predominantly observed in GC tissues and was weaker or no signal was observed in the paired adjacent noncancerous mucosal tissue samples (). These data are consistent with our previous PCR results suggesting that circRNA0047905 was dominantly expressed in GC tissues and may serve as a biomarker for GC in future studies.
Discussion
CircRNAs are generally expressed at low levels and often exhibit cell type-specific and tissue-specific patterns. The functions of most circRNAs remain largely unexplored; nevertheless, known functions include sequestration of microRNAs or proteins, modulation of transcription and interference with splicing, and even translation to produce polypeptides [Citation20]. Several lines of evidence suggest that circRNAs may play key regulatory roles in cancer [Citation21]. In previous studies, we identified a novel circular RNA, called circRNA0047905, that was significantly upregulated in human GC and promoted cell proliferation and migration [Citation10]. In the current study, we present findings that complement this previous study. We constructed a circRNA-miRNA-mRNA network with bioinformatics [Citation12]. This ceRNA network indicated potential associations between circRNAs and their target genes. The networks provided an important reference value for studying the interaction of other differentially expressed circRNAs and their potential targets.
CircRNA0047905 is derived from exon 4 to exon 7 of the SERPINB5 gene. In previous studies, we found that SERPINB5 RNA was also expressed more abundantly in GC than in matched normal tissues, and the expression levels of circRNA0047905 and SERPINB5 RNA were positively correlated [Citation10]. In addition, the frequency of SERPINB5 induction was associated with the stage of gastric cancer (GC) and lymph node metastasis [Citation14,Citation17]. Mechanistically, circRNA0047905 functions as a ceRNA through harboring miR4516 to abolish the suppressive effect on target gene SERPINB5 ().
Figure 9. The schematic diagram shows the mechanism underlying circRNA0047905 as a ceRNA for miR4516 and miR1227-5p.
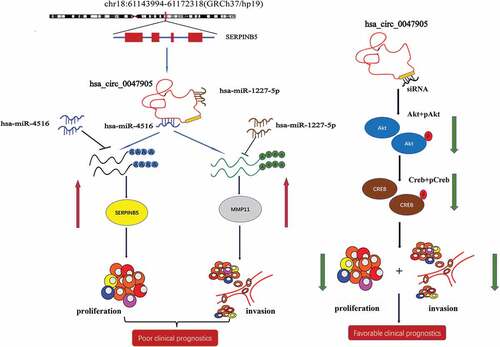
MMP11 is a member of the matrix metalloproteinase (MMP) family, involved in the breakdown of extracellular matrix in normal physiological processes as well as in diseases. Studies have shown that MMP11 is closely related to the proliferation, invasion, lymph node metastasis and prognosis of gastric cancer, suggesting that MMP11 gene also plays an important role in the occurrence and development of gastric cancer [Citation15,Citation22]. We found that MMP11 RNA was also more abundantly expressed in GC than in matched normal tissues. We also found that overexpression of MMP11 correlated with poor prognosis in GC patients [Citation17]. Mechanistically, circRNA0047905 functioned as a ceRNA through harboring miR1227-5p to abolish the suppressive effect on target gene MMP11 ().
Because of the low sensitivity and specificity of the existing biomarkers of gastric cancer, a panel approach of combinations allow us to pursue more precise diagnoses. RNA scope assay was performed in five GC tissue samples. Preliminary results indicated that positive signals were predominantly observed in GC tissues. In order to comprehensively evaluate the potential of circRNA0047905 as a molecular diagnostic marker for gastric cancer, it is necessary to further expand the number of clinical samples on the basis of previous work to detect the expression of circRNA0047905 in gastric cancer tissue samples and blood samples. Our data further indicated that circRNA0047905 may affect the Akt/CERB signaling pathway in gastric cancer progression through molecular mechanisms that are currently unknown, suggesting the need for further detailed exploration in the future ().
Conclusion
Circular RNA profiling of GC tissues allowed identification of a differentially expressed circRNA derived from the SERPINB5 gene. We demonstrated that circRNA0047905 is a proliferative factor and potential biomarker in gastric cancer. Our findings suggest that circRNA0047905 may play important roles in the pathogenesis and development of GC.
Materials and methods
Patient samples
Thirty-one GC tissues and paired adjacent noncancerous mucosal tissue samples were obtained from the Department of General Surgery, Peking University Peoples’ Hospital. The samples were immediately snap-frozen in liquid nitrogen after resection and were stored at −80°C until RNA extraction. The diagnosis of GC was confirmed pathologically. Five pairs of samples were used for microarray analysis, and thirty-one pairs were used for qRT-PCR validation. All patients provided written informed consent prior to sample collection. The study was approved by the local Research Ethics Committee of Peking University.
Cell lines and cell culture
The 293T cell and human GC cell line AGS were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). These cell lines were tested and authenticated for genotypes by DNA fingerprinting. The cell line was passaged for fewer than 6 months after reconstitution. AGS cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (all from Thermo Fisher, USA) at 37°C with 5% CO2. 293T cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (all from Thermo Fisher, USA) at 37°C with 5% CO2.
RNA extraction and purification
Total RNA was extracted from tissue samples using TRIzol reagent (Invitrogen, USA) and were purified using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocol. The purity and concentration of RNA were determined from OD260/280 readings using a spectrophotometer (Nano-Drop ND-1000). RNA integrity was determined by 1% formaldehyde denaturing gel electrophoresis.
CircRNA and mRNA microarray expression profiling
Microarray profiling was conducted in the laboratory of CapitalBio Technology Corporation in Beijing, China. Sample labeling, microarray hybridization and washing were performed according to manufacturer’s standard protocols.
CeRNAs network
Based on the results of aberrantly-expressed RNAs obtained from the previous experimental data, TargetScan and miRanda databases were used to investigate the potential target of miRNAs, and a ceRNA network was constructed according to the common miRNAs binding circRNAs and mRNAs [Citation12]. Through merging the common targeted miRNAs, we constructed a network under the support of bioinformatics analysis with their NovelBio Cloud Analysis Platform.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Reverse transcription was performed using a reverse transcription kit (Invitrogen, USA). Expression levels of circRNAs and mRNAs were measured by qRT-PCR using the iTaq™ Universal SYBR® Green Supermix (Bio-Rad, USA) in the CFX96 Real-Time PCT Detection System (Bio-Rad, Hercules, CA). Human β-Actin RNA was amplified as an internal control for circRNAs and mRNAs. CircRNA and mRNA expression levels were calculated according to the 2−△△Ct method.
Generation of knockdown cell lines
Small interfering RNAs (siRNAs) were synthesized by Ribo Bio Technology Co Ltd (Guangzhou, China) to interfere with the expression of specific genes with complementary nucleotide sequences by degrading circRNAs. siRNAs were transfected with Lipofectamine™ RNAiMAX Transfection Reagent (Invitrogen, USA). The expression of circRNAs and mRNAs in transfected cells were confirmed by qRT-PCR.
Western blot analysis
Total protein from the cells was extracted using cell lysis buffer with proteinase inhibitors (Roche Diagnostics, Basel, Switzerland). Lysates were denatured with sodium dodecyl sulfate (SDS) buffer at 100°C for 10 min, separated in 10% or 15% polyacrylamide gels and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Hertfordshire, UK). The membranes were blocked with 5% non-fat milk powder in Tris-buffered saline containing 0.1% Tween 20 (TBST) and probed with MMP11 antibody (ab53143; Abcam plc, Cambridge, UK), AKT1/2/3 antibody (ab179463; Abcam plc, Cambridge, UK), AKT1 (phospho S473) antibody (ab81283; Abcam plc, Cambridge, UK), CREB antibody (ab31387; Abcam plc, Cambridge, UK) and CREB (phospho S133) antibody (ab32096; Abcam plc, Cambridge, UK) overnight at 4°C. The following day, the membranes were treated with secondary antibodies. The band signals were visualized through enhanced chemiluminescence (ECL; Pierce, Rockford, IL, USA) after exposure to a ChemiDocTM XRSC system (Bio-Rad).
Phosphoprotein profiling by microarray
To further reveal the mechanisms of circRNA0047905 influencing gastric cancer proliferation, we generated a Cancer Signaling Phosphoantibody microarray CSP100 to measure changes in protein expression levels in 12 tumor-related classical signaling pathways (Full Moon Biosystems, Inc., Sunnyvale, CA). Each of the antibodies has two replicates that are printed on a coated glass microscope slide, along with several positive and negative controls. The antibody array experiment was performed by Wayen Biotechnology (Shanghai, China) according to their established protocol. The fluorescence signal of each antibody was obtained from the fluorescence intensity of this antibody spot. A ratio computation was used to measure the extent of protein phosphorylation, calculated as follows: phosphorylation ratio = phospho value/unphospho value.
CircRNA0047905 RNA scope in situ hybridization assay
CircRNA0047905 expression was examined in five paired tissues of GC by RNA in situ hybridization using RNAscope technology (Advanced Cell Diagnostics, Hayward, CA, USA). The probe targeting circRNA0047905 was designed by Advanced Cell Diagnostics Company (USA) and used in the RNAscope assay was provided by Advanced Cell Diagnostics. circRNA0047905 molecules were detected with single-copy detection sensitivity. Single-molecule signals were quantified on a cell-by-cell basis by manual counting. A probe that targeted human PPIB mRNA (Advanced Cell Diagnostics; Cat No.476701) was used as the positive control. A probe targeting Bacillus subtilis DapB mRNA (Advanced Cell Diagnostics; Cat No. 310043) was used as the negative control. The RNA scope in situ hybridization experiment was performed by OUTDO BIOTECH (Shanghai Xinchao Biotechnology Co., LTD, China) according to their established protocol.
Statistical analysis
All results were expressed as means ± SD. Differences between groups were assessed using a paired t-test. FDR<0.05 or P < 0.05 was considered statistically significant. The relationship between the expression of circRNAs and mRNAs was evaluated by Spearman’s correlation. P < 0.05 was considered statistically significant. All data, unless otherwise explained specifically, were analyzed using SPSS 20.0 software (SPSS, Chicago, IL, USA).
Supplemental Material
Download MS Word (378 KB)Acknowledgments
We thank members of our laboratory for helpful discussion and Novel Bioinformatics Ltd., Co for the support of bioinformatics analysis with their NovelBio Cloud Analysis Platform.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132.
- Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977–1010.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358.
- Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148.
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. Rna Biol. 2017;14:514–521.
- Pan RY, Liu P, Zhou HT, et al. Circular RNAs promote TRPM3 expression by inhibiting hsa-miR-130a-3p in coronary artery disease patients. Oncotarget. 2017;8:60280–60290.
- Wang L, Shen J, Jiang Y. Circ_0027599/PHDLA1 suppresses gastric cancer progression by sponging miR-101-3p.1. Cell Biosci. 2018;8:58.
- Lai Z, Yang Y, Yan Y, et al. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. 2017;16:2301–2311.
- Sun TT, He J, Liang Q, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6:784–801.
- Liu Q, Zhang X, Hu X, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation. Sci Rep. 2016;6:22572.
- Song SY, Son HJ, Kim MH, et al. Prognostic significance of maspin expression in human gastric adenocarcinoma. Hepatogastroenterology. 2007;54:973–976.
- Lei KF, Liu BY, Wang YF, et al. SerpinB5 interacts with KHDRBS3 and FBXO32 in gastric cancer cells. Oncol Rep. 2011;26:1115–1120.
- Xu J, Yao Y, Ren S, et al. Matrix metalloproteinase expression and molecular interaction network analysis in gastric cancer. Oncol Lett. 2016;12:2403–2408.
- Deng H, Guo RF, Li WM, et al. Matrix metalloproteinase 11 depletion inhibits cell proliferation in gastric cancer cells. Biochem Biophys Res Commun. 2005;326:274–281.
- Szasz AM, Lanczky A, Nagy A, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333.
- Tan J, Yang L. Long noncoding RNA VPS9D1-AS1 overexpression predicts a poor prognosis in non-small cell lung cancer. Biomed Pharmacother. 2018;106:1600–1606.
- Tamma R, Annese T, Ruggieri S, et al. VEGFA and VEGFR2 RNAscope determination in gastric cancer. J Mol Histol. 20182018;49:429–435.
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442.
- Chen Y, Li C, Tan C, et al. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53:359–365.
- Kou YB, Zhang SY, Zhao BL, et al. Knockdown of MMP11 inhibits proliferation and invasion of gastric cancer cells. Int J Immunopathol Pharmacol. 2013;26:361–370.