ABSTRACT
The spliceosome is a complex molecular machine assembled from many components, which catalyzes the removal of introns from mRNA precursors. Our previous study revealed that the Nrl1 (NRDE-2 like 1) protein associates with spliceosome proteins and regulates pre-mRNA splicing and homologous recombination-dependent R-loop formation in the fission yeast Schizosaccharomyces pombe. Here, we identify proteins associated with splicing factors Ntr1, Ntr2, Brr2 and Gpl1, a poorly characterized G-patch domain-containing protein required for efficient splicing. This work provides new evidence that Nrl1 and splicing factors physically interact and reveals additional insights into the protein interaction network of the spliceosome. We discuss implications of these findings in the light of recent progress in our understanding of how Nrl1 and splicing factors ensure genome stability.
Introduction
Splicing of precursor messenger RNA is catalyzed by the spliceosome, which is assembled from many components around a pre-mRNA substrate. The major components of the spliceosome include small nuclear ribonucleoprotein particles containing U1, U2, U4, U5 or U6 snRNA (snRNPs), the nineteen complex (NTC) and the nineteen-related complex (NTR). In addition, there are several other enzymes and cofactors associated with the spliceosome. Recent structural studies together with previous genetic and biochemical analyses provided important insights into the mechanisms of spliceosome assembly and disassembly as well as activation and catalytic reactions. These studies revealed that the spliceosome shows a remarkable structural and mechanistic conservation between yeast and human systems suggesting that the underlying mechanism is conserved across most eukaryotes [Citation1,Citation2].
Previous studies identified Nrl1, a protein sharing similarities with NRDE-2 proteins, as a spliceosome-associated protein that affects mRNA splicing of a subset of genes and non-coding RNAs. Nrl1 also suppresses homologous recombination-dependent R-loop formation and targets transcripts with cryptic introns to form heterochromatin domains at developmental genes and retrotransposons in the fission yeast Schizosaccharomyces pombe [Citation3,Citation4]. In this Extra View, we provide an analysis of the spliceosome complex and validate the specificity of interactions between Nrl1 and spliceosome by purifying TAP-tagged splicing factors Ntr1, Ntr2, Brr2 and SPAC20H4.06c, a G-patch domain-containing protein orthologous to human GPATCH1 protein, which we named Gpl1 (GPATCH1 like 1) [Citation5,Citation6]. We also discuss insights into the protein interaction network of splicing factors including a possible functional interaction between G-patch domain-containing protein Gpl1 and putative RNA helicase SPAC20H4.09.
Results and discussion
In our previous study, we used tandem affinity purification (TAP) protocol followed by mass-spectrometry analysis to identify proteins associated with Nrl1. We found that splicing factors Ntr1, Ntr2, Brr2 and Gpl1, a poorly characterized G-patch domain-containing protein required for efficient splicing, as well as several other proteins co-purified with Nrl1-TAP [Citation3]. An independent study found that Ntr2 and Gpl1 as well as other splicing factors co-purified with Nrl1-MYC and Nrl1-FLAG [Citation4]. Moreover, we observed an interaction between Nrl1 and Ntr2 in a yeast two-hybrid assay [Citation3].
In order to validate the specificity of interactions between Nrl1 and spliceosomal factors, we decided to perform reciprocal purifications using TAP-tagged Ntr1, Ntr2, Brr2 and Gpl1. We constructed strains expressing TAP-tagged Ntr1 (Ntr1-TAP), Ntr2 (Ntr2-TAP), Brr2 (Brr2-TAP) and Gpl1 (Gpl1-TAP), according to our protocol described at http://mendel.imp.ac.at/Pombe_tagging/ [7]. While ntr1, ntr2 and brr2 genes are essential for cell growth [Citation8,Citation9], haploid cells expressing Ntr1-TAP, Ntr2-TAP or Brr2-TAP were viable (data not shown). We used a TAP protocol to purify Ntr1-TAP, Ntr2-TAP, Brr2-TAP and Gpl1-TAP from cycling S. pombe cells and identified co-purifying proteins by mass spectrometry (,)). Indeed, we found that Nrl1 co-purified with Ntr11-TAP, Ntr2-TAP, Brr2-TAP as well as Gpl1-TAP (), Table S1). In addition to Nrl1, we identified many of the known components of the spliceosome with high scores in our purifications (), Table S1), suggesting that our purifications were specific and many important interactions were preserved.
Figure 1. Identification of proteins co-purifying with S. pombe Ntr1-TAP, Ntr2-TAP, Brr2-TAP and Gpl1-TAP.
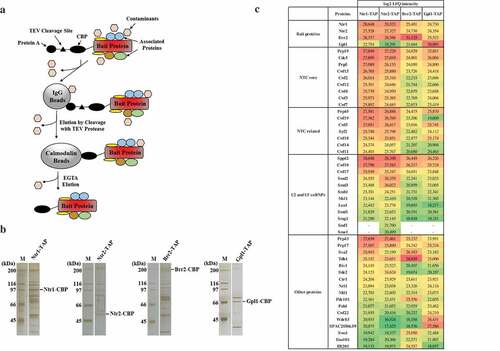
While many splicing factors such as the Brr2 helicase have been studied in great detail, other proteins involved in splicing are poorly characterized. Gpl1 is a G-patch domain-containing protein required for efficient splicing, but its molecular function is not known [Citation10]. G-patch domain has a characteristic glycine-rich sequence signature, and it is present in many eukaryotic proteins involved in RNA-processing [Citation11]. Previous studies showed that G-patch domain proteins contribute to the function of RNA helicases involved in RNA processing [Citation10,Citation12,Citation13]. Interestingly, we found that Gpl1-TAP co-purified with SPAC20H4.09 (), Table S1), an uncharacterized RNA helicase that shares similarities with human DHX35 [Citation14]. Previous observations that both Gpl1 and SPAC20H4.09 co-purify with splicing factors [Citation15–Citation17] and that SPAC20H4.09Δ mutant shows genetic interactions with mutants defective in splicing [Citation18] are consistent with the possible role of SPAC20H4.09 in RNA splicing. SPAC20H4.09 and gpl1 genes are located on the same chromosome, separated by only about 4000 bp, suggesting possible common regulation of their expression [Citation14]. Therefore, we speculate that the splicing defect observed in gpl1Δ mutant cells might be due to impaired function of the SPAC20H4.09 helicase. Further functional studies of the SPAC20H4.09 helicase and its possible regulation by the G-patch protein Gpl1 are needed to understand their roles in RNA splicing.
Taken together, we show that Nrl1 co-purifies with Ntr1-TAP, Ntr2-TAP, Brr2-TAP and Gpl1-TAP, confirming our previous observation that Nrl1 interacts with splicing factors. In addition, our results provide new insights into the protein interaction network of splicing factors. Based on our results, we speculate that a putative RNA helicase SPAC20H4.09 functionally interacts with the G-patch domain-containing protein Gpl1 required for efficient splicing. Further exploration should reveal a better understanding of the functional relationship between proteins identified in our purifications. Given the conserved nature of these proteins, we expect that our work will shed light on the functions of these proteins both in yeast and higher eukaryotes.
Materials and methods
Strains
S. pombe strains expressing either Ntr1-TAP (FY310, h− leu1-32 ura4-D18 ade6-M216 ntr1-TAP::KanMX6), Ntr2-TAP (FY311, h− leu1-32 ura4-D18 ade6-M216 ntr2-TAP::KanMX6), Brr2-TAP (FY307, h− leu1-32 ura4-D18 ade6-M216 brr2-TAP::KanMX6) or Gpl1-TAP (FY331, h− leu1-32 ura4-D18 ade6-M216 gpl1-TAP::KanMX6) were grown in complete yeast extract medium (5.0 g/l yeast extract, 3.0% glucose, 0.1 g/l L-leucine, 0.1 g/l L-lysine hydrochloride, 0.1 g/l L-histidine, 0.1 g/l uracil and 0.15 g/l adenine sulfate) at 32°C. TAP-tagging was confirmed by PCR and immunoblotting using PAP antibody (rabbit antiperoxidase antibody linked to peroxidase, Dako) at 1:20.000 dilution (2% skim milk in 0.05% PBS-T).
Protein purification
Fifteen-liter cultures of strains expressing TAP-tagged proteins were grown to mid-log phase (OD600 ~ 0.8–1.0) and cells were collected by centrifugation. Yeast cell powders (50 g) were made from frozen cell pellets using SPEX SamplePrep 6770 Freezer/Mill cooled by liquid nitrogen. Proteins were extracted using IPP150 buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% NP-40, complete protease and phosphatase inhibitors and 1 mM PMSF). Five hundred microliters of IgG Sepharose™ 6 Fast Flow beads per sample (GE Healthcare) was washed with IPP150 buffer, mixed with protein extracts and rotated for 2 h at 4°C. Beads were washed with 20 volumes of IPP150 buffer and with 5 volumes of TEV cleavage buffer (TCB, 10 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% NP-40, 0.5 mM EDTA and 1 mM DTT). Cleavage step was performed in 2 ml of TCB buffer supplemented with 400 Units of Turbo TEV protease (MoBiTec) for 2 h at 16°C. Two milliliters of eluates was supplemented with 6 µl of 1 M CaCl2 and mixed with 6 ml of Calmodulin binding buffer 1 (CBB1, 10 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% NP-40, 1 mM imidazole, 1 mM Mg-Acetate, 2 mM CaCl2 and 10 mM β-mercaptoethanol). One hundred and fifty microliters of Calmodulin Sepharose™ 4B beads per sample (GE Healthcare) was washed with CBB1 buffer, added to a mixture of eluates and CBB1 buffer and incubated for 2 h at 4°C. The beads were washed with 10 volumes of CBB1 and 5 volumes of Calmodulin binding buffer 2 (CBB2, 10 mM Tris pH 8.0, 150 mM NaCl, 1 mM Mg-Acetate, 2 mM CaCl2 and 1 mM β-mercaptoethanol). The proteins were step-eluted using bead volume of elution buffer (EB, 10 mM Tris pH 8.0, 150 mM NaCl, 1 mM Mg-acetate, 2 mM EGTA and 1 mM β-mercaptoethanol). Eluted proteins were separated on SDS-PAGE and visualized by silver staining. Eluates from peak fractions were submitted for LC-MS/MS analysis.
LC-MS/MS analysis
Samples were reduced in the presence of 10 mM DTT (30 min, 60°C), alkylated by addition of 30 mM iodoacetamide (20 min, RT/in dark) and the alkylation reaction was quenched by additional 10 mM DTT. One microgram of modified sequencing grade trypsin (Promega) was added to the protein mixture, and the samples were incubated overnight at 37°C. The reaction mixture was acidified by addition of 0.5% TFA, the peptides were purified by microtip C18 SPE and dried in the speedVac. For LC-MS analysis, the set of nanotrap column (Acclaim PepMap100 C18, 75 μm x 20 mm, Dionex, CA, USA) and nanoseparation column (Acclaim PepMap C18, 75 μm x 500 mm, Dionex) attached to UltiMate 3000 RSLCnano system (Dionex, CA, USA) were used. The peptides were separated in 1 h gradient from 3% to 43% B with two mobile phases used: 0.1% FA (v/v) (A) and 80% ACN (v/v) with 0.1% FA (B). Spectral datasets were collected by Orbitrap Elite mass spectrometer (ThermoScientific, MA, USA) operating in the data-dependent mode using Top15 strategy for the selection of precursor ions for the HCD fragmentation [Citation19]. Each of the samples was analyzed in two technical replicates. Obtained datasets were processed by MaxQuant (version 1.5.3.30) [Citation20] with built-in Andromeda search engine using carbamidomethylation (C) as permanent modification and oxidation (M), acetylation (N-terminus) and phosphorylation (STY) as variable modifications. The analysis was performed as LFQ experiment using “match between runs” as an advanced parameter [Citation21]. The search was performed against the S. pombe protein database (UniProt, downloaded 25. 08. 2016).
Supplemental Material
Download Zip (470.4 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Plaschka C, Newman AJ, Nagai K. Structural basis of nuclear pre-mRNA splicing: lessons from yeast. Cold Spring Harb Perspect Biol. 2019;11:a032391.
- Yan C, Wan R, Shi Y. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Cold Spring Harb Perspect Biol. 2019;11:a032409.
- Aronica L, Kasparek T, Ruchman D, et al. The spliceosome-associated protein Nrl1 suppresses homologous recombination-dependent R-loop formation in fission yeast. Nucleic Acids Res. 2016;44:1703–1717.
- Lee NN, Chalamcharla VR, Reyes-Turcu F, et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013;155:1061–1074.
- Absmeier E, Santos KF, Wahl MC. Functions and regulation of the Brr2 RNA helicase during splicing. Cell Cycle. 2016;15:3362–3377.
- Fair BJ, Pleiss JA. The power of fission: yeast as a tool for understanding complex splicing. Curr Genet. 2017;63:375–380.
- Cipak L, Spirek M, Novatchkova M, et al. An improved strategy for tandem affinity purification-tagging of Schizosaccharomyces pombe genes. Proteomics. 2009;9:4825–4828.
- Kim DU, Hayles J, Kim D, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623.
- Spirek M, Benko Z, Carnecka M, et al. S. pombe genome deletion project: an update. Cell Cycle. 2010;9:2399–2402.
- Larson A, Fair BJ, Pleiss JA. Interconnections between RNA-processing pathways revealed by a sequencing-based genetic screen for pre-mRNA splicing mutants in fission yeast. G3 (Bethesda). 2016;6:1513–1523.
- Aravind L, Koonin EV. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem Sci. 1999;24:342–344.
- Robert-Paganin J, Halladjian M, Blaud M, et al. Functional link between DEAH/RHA helicase Prp43 activation and ATP base binding. Nucleic Acids Res. 2017;45:1539–1552.
- Warkocki Z, Schneider C, Mozaffari-Jovin S, et al. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 2015;29:94–107.
- Lock A, Rutherford K, Harris MA, et al. PomBase 2018: user-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res. 2019;47:D821–D827.
- Thakran P, Pandit PA, Datta S, et al. Sde2 is an intron-specific pre-mRNA splicing regulator activated by ubiquitin-like processing. Embo J. 2018;37:89–101.
- Ren L, McLean JR, Hazbun TR, et al. Systematic two-hybrid and comparative proteomic analyses reveal novel yeast pre-mRNA splicing factors connected to Prp19. PLoS One. 2011;6:e16719.
- Yan C, Hang J, Wan R, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191.
- Ryan CJ, Roguev A, Patrick K, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46:691–704.
- Michalski A, Damoc E, Lange O, et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics. 2012;11:O111 013698.
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372.
- Cox J, Hein MY, Luber CA, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526.
