ABSTRACT
LncRNA DANCR has been proven to be involved in osteoblast differentiation. This study aims to investigate the role of DANCR in osteoclast formation and root resorption in periodontal ligament (PDL) cells induced by compression force (CF). Rat orthodontic tooth movement (OTM) model was established. The molecules expressions in the areas of root resorption form OTM model were measured. The number of osteoclasts was measured using Tartrate-resistant acid phosphatase (TRAP) staining. The bone resorption was detected using pit formation assay. We showed that the expression of DANCR and Jagged1 protein was increased in rat OTM model and human periodontal ligament (hPDL) cells treated with CF, and CF increased the production of Jagged1, RANKL, and IL-6 from the hPDL cells. Moreover, DANCR could positively regulate Jagged1 protein expression. Knockdown of DANCR could change the promotion effect of CF on osteoclastogenesis and bone resorption in vitro and in vivo experiments, while overexpression of Jagged1 reversed si-DANCR effect. Taken together, knockdown of DANCR reduced osteoclast formation and root resorption induced by CF via Jagged1.
1 Introduction
Orthodontic treatment is becoming more and more popular among adults [Citation1]. During the course of orthodontic treatment, periodontal tissue is reconstructed under the induction of orthodontic force. Orthodontically induced inflammatory root resorption (OIIRR) is an unavoidable pathologic consequence of orthodontic treatment, which can cause permanent loss of the dental structure of the root apex [Citation2,Citation3].
Notch signaling pathway plays an important role in osteoclast maturation and resorptive activity [Citation4]. Jagged1 is one of five Notch ligands, which has been first identified in 1995 [Citation5]. Previous study showed that Jagged1 was increased in the areas of root resorption during experimental tooth movement in rats in vivo [Citation3]. Moreover, Nakao et al. [Citation6] reported that parathyroid hormone related protein promoted receptor activator of nuclear factor κB ligand (RANKL)-induced osteoclastogenesis through increasing Jagged1, thus likely promoting tooth resorption. Therefore, Jagged1 may contribute to OIIRR. However, little is known about the mechanism of Jagged1 in OIIRR.
Long non-coding RNA (lncRNA), a class of RNA with the length more than 200 nt, can play crucial regulatory roles in protein modification, transcriptional level, and regulation of epigenetic level [Citation7]. Recently, lncRNA has been found to be involved in bone development, e.g. lncRNA LINC00311 could promote the proliferation and differentiation of osteoclasts in osteoporotic Rats by targeting DLL3 [Citation8]; lncRNA AK077216 contributed to RANKL-induced osteoclastogenesis and bone resorption via NFATc1/NIP45 [Citation9]. LncRNA DANCR (differentiation antagonizing non-protein coding RNA) is an oncogene in various tumors, such as prostate cancer [Citation10] glioma [Citation11], gastric cancer [Citation12], etc. Recent studies have shown that DANCR is involved in osteoblast differentiation. Zhang et al. [Citation13] revealed that knockdown of DANCR promoted the osteogenic differentiation of the HBMSCs via the p38 MAPK pathway. Tang et al. [Citation14] showed that DANCR could inhibit osteoblast differentiation via regulating FOXO1, and then participate in osteolysis after total hip arthroplasty. These findings suggest that DANCR is involved in bone development, but the relationship between DANCR and OIIRR has not been reported. Based on the target gene prediction database (LncBase Predicted v.2), DANCR is one of the binding target lncRNAs of miR-34a-5p, which has been considered as an inhibitor for osteoclastogenesis [Citation15]. In addition, miR-34a-5p is also related to orthodontic treatment. It can enhance alveolar bone remodeling under orthodontic force in vivo [Citation16].
Therefore, the aims of this study were to determine the expression of miR-34a-5p, DANCR and Jagged1 in the areas of root resorption during experimental tooth movement in rats in vivo and in human periodontal ligament (hPDL) cells subjected to compression force (CF), and investigate the effects of DANCR/Jagged1 on hPDL cells-mediated osteoclastogenesis and bone resorption.
2 Methods
2.1 Animals
Wistar rats (body weight 190 ± 15 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The animal experimental protocol used in this study was approved by the Ethics Committee for Animal Experiments at the First Affiliated Hospital of Zhengzhou University.
2.2 Rat orthodontic tooth movement model
Rats were anesthetized using pentobarbital sodium (40 mg/kg body weight). The rat orthodontic tooth movement (OTM) model was established as previously described [Citation2]. A 0.008 inch stainless steel ligature wire was used to ligate a closed-coil spring (0.005 inch in wire size, 0.083 inch in diameter; Grikin Advanced Materials Co. Ltd, Beijing, China) to the maxillary first molar cleat. The other side of the closed-coil spring was also ligated using the same stainless steel ligature wire, with the holes in the maxillary incisors drilled laterally just above the gingival papilla with a #1/4 round bur. The upper first molar was moved mesially by the closed coil spring with a CF of 50 g.
Rats were divided into 4 groups. (1) Control group: rats did not receive any CF. (2) OTM group: the OTM model group. (3) si-DANCR group. (4) si-control group. si-DANCR or si-control (1 nmol/time, 50 μl) was local injected into the buccal and lingual submucous between the first and second molars for three consecutive days. Then, rats were subjected to tooth movement. Each group had six rats. Seven days after modeling, PDL tissues of each rat were obtained for tartrate-resistant acid phosphatase (TRAP) staining, qRT-PCR and Western blot.
2.3 Immunohistochemical staining
Rats were anesthetized with thiamylal sodium and myocardial perfusion with 4% paraformaldehyde solution in 0.1 M phosphate buffer (pH 7.4). Then, the maxilla was immediately dissected and fixed with the same fixative at 4°C overnight. The specimens were decalcified by 10% EDTA (pH 7.4) for 5 w. Next, the decalcified specimens were embedded in paraffin after dehydration in a graded ethanol series. The samples were sliced into 4 µm sections and stained using TRAP staining Kit (MX80187.3, Ming Xiu (Shanghai) Biotechnology Co., Ltd., Shanghai, China). Five fields (×400) were randomly selected, and the number of TRAP-positive osteoclasts in each field was counted. The average value was calculated.
2.4 Human PDL cell culture
Human periodontal ligament (hPDL) cells were prepared according to the method of Nakajima et al. [Citation17]. Briefly, hPDL tissues were taken from the roots of premolars extracted from six healthy young volunteers (three males and three females; 14‒16 y old) during the course of orthodontic treatment. The study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University. All donors signed the informed consent form. The periodontal ligament tissues were placed in 35-mm tissue culture dishes and covered with a sterilized glass coverslip. The medium was α-minimal essential medium (Gibco, NY, USA), which was supplemented with 100 µg/mL of penicillin-G (Sigma, MO, USA), 50 µg/mL of gentamicin sulfate (Sigma), 0.3 µg/mL of amphotericin B (Flow Laboratories, VA, USA), and 10% fetal calf serum (Cell Culture Laboratories, OH, USA). The cultures were kept at 37°C in a humidified incubator with 95% air and 5% CO2. When the cells growing from each explant reached confluence, they were detached with 0.05% trypsin in PBS for 10 min and subcultured in culture flasks. The cells still adhered to the bottom of the flask were discarded to avoid contamination by epithelial cells.
2.5 Application of CF
To reproduce the pressure conditions during the OTM, we performed the following in vitro experiments as previously described [Citation3]. The hPDL cells were continuously compressed using a uniform compression system as a model of the pressure at the site of orthodontic movement. The cells were subsequently subjected to 4.0 g/cm2 of CF for 1, 3, 6, 9, 12, and 24 h.
2.6 Human osteoclast precursor cell culture
Human osteoclast precursor cells (hOCPs, cell line: RAW 264.7 cells) were purchased from Cell bank of the Chinese Academy of Sciences, Shanghai, China. The following in vitro experiments were conducted as previously described [Citation3].
For TRAP staining, hOCPs (1 × 104 cells/100 µL) were seeded onto 16-well Nunc Lab-Tek chamber slides (ThermoFisher Scientific, Waltham, MA) and cultured in commercial medium including the conditioned medium at 37°C for five days. Then, the osteoclasts were washed twice with PBS and stained using a TRAP staining Kit (MX80187.1, Ming Xiu (Shanghai) Biotechnology Co., Ltd.). The conditioned medium was the culture medium of hPDL cells that treated with CF, si-DANCR, or Lenti-Jagged1-GFP for 24 h.
For the pit formation assay, hOCPs were seeded onto 16-well Nunc Lab-Tek chamber slides and cultured in commercial medium for seven days. After the osteoclasts matured, the commercial medium was changed to the conditioned medium (same as above), and the cells were incubated for another 72 h.
2.7 Cell transfection
MiR-34a-5p inhibitor, miR-34a-5p mimic, pcDNA-DANCR, and si-DANCR were synthesized by GenePharma (Shanghai, China). For transfection, the miR-34a-5p mimic, mimic control, the miR-34a-5p inhibitor, inhibitor control, pcDNA-DANCR, or si-DANCR were diluted at a final concentration of 100 nM. Briefly, hPDL cells were seeded into 24-well plates at a density of 2 × 105 cells/well. When the cultures reached 70‒80% confluence, the cell transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instruction. 24 h following transfection, cells were harvested for qRT-PCR and western blot analyses.
2.8 Enzyme-linked immunosorbent assay
The conditioned medium was collected for Enzyme-linked immunosorbent assay (ELISA). The levels of Jagged1, RANKL and IL-6 released into the culture supernatants were measured using Jagged1 ELISA Kit (Cusabio, Wuhan, China), RANKL ELISA Kit (Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China), and IL-6 ELISA Kit (Abcam, USA), respectively.
2.9 Pit formation assay
Pit formation assay was performed according to the method of Asano et al. [Citation2]. Briefly, the bone-resorbing activity of mature osteoclasts formed in vitro was assessed based on the ability of hOCPs to form resorption pits on dentin slices (0.15 mm in diameter, 6 mm in thickness). Scanning electron microscopy (JSM-6510; JEOL, Japan) was used to examine the entire surface of each dentin slice. The resorption area in a single well was determined using an image processing system.
2.10 Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells or tissues using Trizol reagent (Invitrogen, CA, USA). cDNA was then reversely transcribed using PrimeScript® RT reagent Kit (Takara, Shiga, Japan). qRT-PCR reactions were conducted by ABI Prism 5700 Sequence Detection System (Applied Biosystems). The sequence of primers used for amplification is listed: miR-34a-5p (F 5´-CCCACATTTCCTTCTTATCAACAG-3´, R 5´ -GGCATCTCTCGCTTCATCTT-3´); DANCR (F 5´-CTGCATTCCTGAACCGTTATCT-3´, R 5´-GGGTGTAATCCACGTTTCTCAT-3ʹ); U6 (F 5´-TGCGGGTGCTCGCTTCGCAGC-3´, R 5´-CCAGTGCAGGGTCCGAGGT-3´); Jagged1 (F 5´-ATCGTGCTGCCTTTCAGTTT-3´, R 5´GATCATGCCCGAGTGAGAA-3´); β-actin (F 5´-CCTGGCACCCAGCACAAT-3´, R 5´-GGGCCGGACTCGTCATAC-3´). The 2−∆∆CT method was used to calculate the relative expression, with U6 served as the internal control gene of miR-34a-5p, and β-actin ad the internal control gene of DANCR and Jagged1. All measures were repeated three times.
2.11 Western blot
Total protein was extracted from tissues or cells using lysis buffer on ice for 30 min, and quantified with the BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Samples that contained same amount of proteins were then separated by 10% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 5% non-fat milk for 1 h at room temperature, followed by incubation with primary antibodies, including anti-Jagged1 antibody (1/500, ab109536, Abcam, UK), anti-RANKL antibody (1/500, ab9957, Abcam), and anti-β-actin antibody (1/1000, ab8227, Abcam), at 4°C overnight. After that, the membranes were incubated with secondary antibodies (Abcam) at room temperature for 1 h. Proteins were visualized using an enhanced chemiluminescence system (Thermo Fisher Scientific, USA). β-actin was served as a control protein to quantify the expression of related proteins.
2.12 Dual-luciferase reporter assay
DANCR fragments with wild type (WT) or mutant (Mut) miR-34a-5p binding sites were cloned by PCR. The PCR products were inserted into the pmirGLO vector (Promega, USA). Then, the vectors and miR-34a-5p inhibitor (or miR-34a-5p mimic) were transiently co-transfected into HEK-293 cells using Lipofectamine 2000 (Thermo Fisher Scientific). The relative luciferase activity was measured using a luciferase assay kit (Promega) after 48 h transfection.
2.13 RNA immunoprecipitation
The EZ-Magna RIP Kit (Millipore, USA) was used for RNA immunoprecipitation (RIP) experiment to verify the binding between miR-34a-5p and DANCR. HEK-293 cells were transfected with miR-34a-5p mimic (or miR-34a-5p inhibitor) and lysed in lysis buffer, following by incubation with RIP buffer including magnetic beads coupled with anti-Ago2 antibody (Abcam). After immunecoprecipitated RNA was isolated, RT-qPCR was performed to analyze the levels of DANCR and miR-34a-5p in the precipitates.
The RNA-Binding Protein Immunoprecipitation Kit (Millipore) was used to verify the binding between DANCR and Jagged1. HEK-293 cells were lysed in lysis buffer, following by incubation with AGO2 antibody or normal mouse IgG. RNA-protein complexes were immunoprecipitated with protein A agarose beads and RNA was extracted by using Trizol (Invitrogen). The IP-western was used to detect AGO2 and Jagged1 protein level and qRT-PCR was performed to quantify the DANCR.
2.14 RNA pull-down
RNA pull-down assay was performed to verify the interaction between DANCR and Jagged1. The DNA probe complementary to DANCR was synthesized and biotinylated by GenePharma (Shanghai, China). A magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific) was used to conduct RNA pull-down assay according to the manufacturer’s instructions. The RNA-protein binding complexes were identified by western blot.
2.15 Ubiquitination assay
hPDL cells were transfected with HA-Jagged1 (beyotime, Shanghai, China), and si-DANCR (GenePharma) using Lipofectamine 2000 (Invitrogen). After 24 h of transfection, MG132 (10 µg/ml, Sigma, USA) was added into culture medium, and the cells were incubated for another 2 h. Then, cells were washed with PBS for twice, and lysed on ice for 20 min. The cell lysates were immunoprecipitated with the labeled antibodies at 4°C overnight. Western blot was used to detect the eluted proteins.
2.16 Statistical analysis
Data were expressed as mean ± standard deviation, and SPSS 17.0 was used for the data analysis. Comparisons between two groups were completed by Student’s t-test. One-way ANOVA followed by Bonferroni post-hoc test was used to evaluate the statistical significance between different groups. A value of P < 0.05 was considered statistically significant.
3 Results
3.1 DANCR was highly expressed in both in vivo and in vitro OTM model
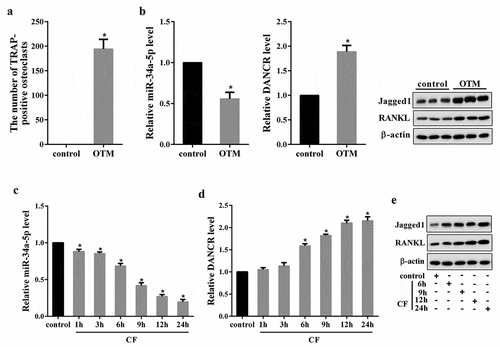
We first tested DANCR expression in OTM rat model. As shown in ), the number of TRAP-positive osteoclasts in OTM group was significantly increased compared with the control. Moreover, miR-34a-5p expression in OTM group was reduced, DANCR expression, as well as the protein expression of Jagged1 and RANKL (the marker molecule for root resorption [Citation18,Citation19]), was up-regulated ()). Then, we measured DANCR expression in hPDL cells treated with CF for 1, 3, 6, 9, 12, and 24 h. Results showed that miR-34a-5p expression in the CF group was significantly decreased following the application of CF in a time dependent manner, reaching the lowest point at 24 h ()). In contrast, DANCR expression and the protein expression of Jagged1 and RANKL was increased following the application of CF in a time-dependent manner, peaking at 24 h (,e)).
Figure 1. The expressions of DANCR, miR-34a-5p, Jagged1 and RANKL in both in vivo and in vitro OTM model. Rats were divided into control and OTM rat model group (n = 6). Seven days after modeling, PDL tissues of each rat were obtained for TRAP staining, qRT-PCR and Western blot. (a) The number of TRAP-positive osteoclasts. (B) The expressions of miR-34a-5p, DANCR, Jagged1 and RANKL in PDL tissues. *P < 0.05 vs control. hPDL cells treated with 4.0 g/cm2 of CF for 1, 3, 6, 9, 12, and 24 h, qRT-PCR and Western blot was used to measure the molecules expressions in the cells.(c) miR-34a-5p expression in hPDL cells. (d) DANCR expression in hPDL cells. (e) The protein expression of Jagged1 and RANKL in hPDL cells. *P < 0.05 vs. control.
3.2 The effect of CF on DANCR expression through miR-34a-5p
Target gene prediction database (LncBase Predicted v.2) predicted a combination of miR-34a-5p and DANCR ()). To confirm this possibility, we performed dual luciferase reporter assay in HEK-293 cells. As shown in ), the luciferase activity of DANCR-WT was enhanced in HEK-293 cells co-transfected with miR-34a-5p inhibitor, while the luciferase activity was reduced in cells co-transfected with miR-34a-5p mimic. The luciferase activity of DANCR-Mut was unaffected in the two groups. In addition, knockdown of miR-34a-5p could up-regulate DANCR expression in hPDL cells, and overexpression of miR-34a-5p could down-regulate DANCR expression in hPDL cells ()). Next, RIP assay was conducted to investigate whether both DANCR and miR-34a-5p might be in the RNA-induced silencing complex, and RNA levels of DANCR and miR-34a-5p in immunoprecipitates were determined by qRT-PCR. We find that DANCR and miR-34a-5p were enriched in Ago2 beads ()).
Figure 2. The effect of CF on DANCR expression through miR-34a-5p. (a) Target gene prediction database (LncBase Predicted v.2) predicted a combination of miR-34a-5p and DANCR. (b) The luciferase activity of DANCR-WT or DANCR-Mut in HEK-293 cells co-transfected with miR-34a-5p inhibitor or miR-34a-5p mimic was measured using dual luciferase reporter assay. (c) Jagged1 expression in hPDL cells transfected with miR-34a-5p inhibitor or miR-34a-5p mimic. (d) The binding between miR-34a-5p and DANCR was measured using RIP assay. *P < 0.05 vs. NC; #P < 0.05 vs. pre-NC. (e) hPDL cells were divided into control, CF, CF+pre-NC, and CF+miR-34a-5p mimic group. MiR-34a-5p expression was measured by qRT-PCR. (f) DANCR expression in hPDL cells. *P < 0.05 vs. control; #P < 0.05 vs. CF+pre-NC.
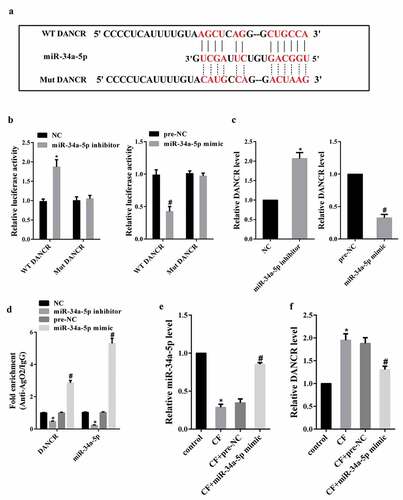
Then, we investigated whether CF affected DANCR expression through miR-34a-5p. hPDL cells were divided into control, CF, CF+pre-NC, and CF+miR-34a-5p mimic group. qRT-PCR analysis showed that CF treatment reduced miR-34a-5p expression, but miR-34a-5p mimic changed this decrease ()). Meanwhile, overexpression of miR-34a-5p could inhibit the increase of DANCR expression induced by CF ()).
3.3 The interaction between DANCR and jagged1
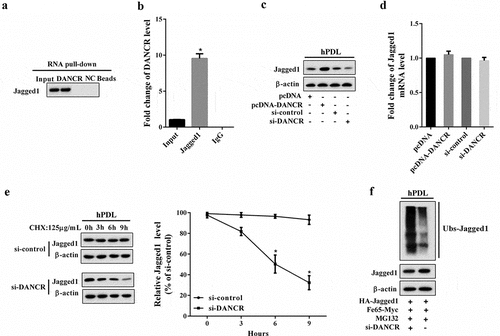
We first performed RNA pull-down assay to explore the interaction between DANCR and Jagged1, and the results revealed that Jagged1 was enriched in the DANCR drop-down complex ()). RIP assay showed that DANCR accumulated in protein samples precipitated by Jagged1 ()). These results suggested that DANCR could interact with Jagged1 in hPDL cells. Then, we investigated how DANCR regulated Jagged1 expression. We find that overexpression of DANCR could increase Jagged1 protein expression, and knockdown of DANCR inhibited Jagged1 protein expression ()). The mRNA expression of Jagged1 was not affected by changes in DANCR expression ()), implying that DANCR might affect the Jagged1 protein stability but its mRNA level. Therefore, we then assessed the effect of DANCR on Jagged1 protein stability via the cycloheximide (CHX, an inhibitor of protein synthesis) assay. hPDL cells were transfected with si-DANCR and treated with 125 μg/mL CHX for 0, 3, 6, and 9 h. As shown in ), interference of DANCR promotes protein degradation of Jagged1 in hPDL cells under CHX treatment, in a time dependent manner. Ubiquitination assay also confirmed that knockdown of DANCR inhibited Jagged1 ubiquitination ()).
Figure 3. The interaction between DANCR and Jagged1. (a) RNA pull-down assay was performed to verify the interaction between DANCR and Jagged1. (b) The binding between DANCR and Jagged1 was measured using RIP assay. *P < 0.05 vs. IgG. (c) hPDL cells were divided into pcDNA, pcDNA-DANCR, si-control, and si-DANCR group. in hPDL cells was measured using Western blot. (d) The mRNA expression of Jagged1 in hPDL cells. (e) After transfection of si-DANCR, hPDL cells were treated with CHX (125 μg/mL) for 0, 3, 6, and 9 h. Western blot was used to detect Jagged1 protein expression. *P < 0.05 vs. control. (f) The effect of DANCR on Jagged1 ubiquitination was confirmed by Ubiquitination assay.
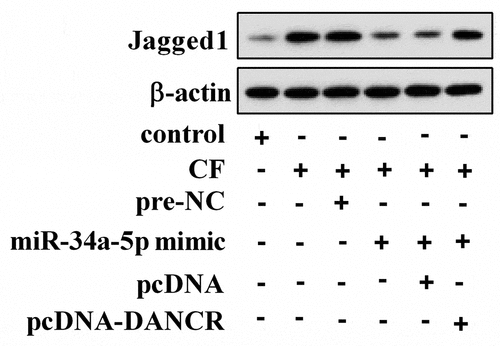
3.4 CF affected jagged1 expression through miR-34a-5p/DANCR
To investigate how CF affected Jagged1 expression, hPDL cells were treated with CF, miR-34a-5p mimic, and pcDNA-DANCR. As shown in , CF treatment could up-regulate Jagged1 protein expression in hPDL cells, but overexpression of miR-34a-5p changed the effect of CF on Jagged1. As expected, overexpression of DANCR transformed the role of miR-34a-5p.
3.5 The effect of CF on osteoclast formation and bone resorption through DANCR/jagged1
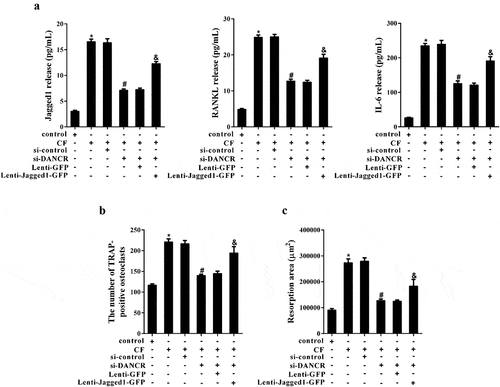
Inflammatory factors produced by hPDL cells treated with CF can stimulate osteoclast activation and cause bone resorption. In the experiments, we first examined the release of inflammatory factors in the supernatant of hPDL cells after different treatments. The results revealed that knockdown of DANCR inhibited the increase of Jagged1, RANKL and IL-6 levels in the supernatant of hPDL cells treated with CF, while overexpression of Jagged1 changed the effect of si-DANCR ()). Then, hPDL cell culture medium from different treatment groups co-cultured with hOCPs respectively. TRAP staining suggested that the number of osteoclasts increased significantly in CF group, and the number reduced obviously in CF+si-DANCR group. As expected, the number was increased in CF+si-DANCR+Lenti-Jagged 1-GFP group ()). The pit formation assay showed that the resorption area in CF group was enhanced, while the resorption area was decreased in CF+si-DANCR group. After transfection of Lenti-Jagged 1-GFP, the resorption area was increased again ()).
Figure 5. The effect of CF on osteoclast formation and bone resorption through DANCR/Jagged1. hPDL cells were divided into control, CF, CF+si-control, CF+si-DANCR, CF+si-DANCR+Lenti-GFP, and CF+si-DANCR+Lenti-Jagged1-GFP group. After 24 h, the supernatant of hPDL cells were collected for ELSA assay. (a) Jagged, RANKL and IL-6 levels in cell supernatant. (b) hOCPs were cultured in hPDL cell culture medium from the above groups for five days. The number of TRAP-positive osteoclasts was measured by TRAP staining. (c) hOCPs were cultured in commercial medium for seven days, and then cultured in hPDL cell culture medium from the above groups. Bone resorption was measured using pit formation assay. *P < 0.05 vs. control; #P < 0.05 vs. CF+si-control; &P < 0.05 vs. CF +si-DANCR + Lenti-GFP.
3.6 Knockdown of DANCR reduced root resorption in rat OTM model
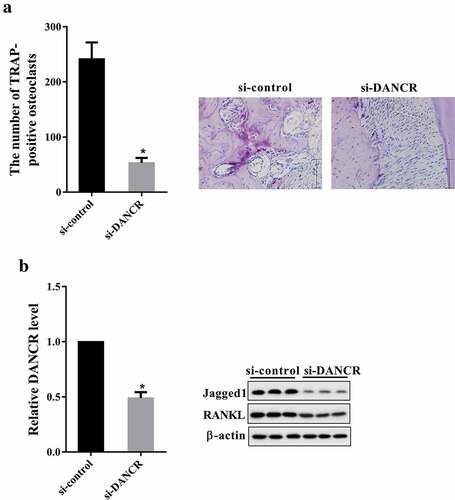
Rats were divided into si-control and si-DANCR group. Seven days after treatment, PDL tissues were collected for TRAP staining, qRT-PCR and western blot. As shown in ), knockdown of DANCR reduced the number of TRAP-positive osteoclasts. The DANCR expression in si-DANCR group was less than in si-control group ()). Moreover, the protein expression of Jagged1 and RANKL was down-regulated in si-DANCR group compared with si-control group ()).
Figure 6. Knockdown of DANCR reduced root resorption in rat OTM model. Rats were divided into si-control and si-DANCR group. Each group had six rats. si-DANCR or si-control (1 nmol/time, 50 μl) was local injected into the buccal and lingual submucous between the first and second molars for three consecutive days. After seven days, PDL tissues of each rat were obtained for for TRAP staining, qRT-PCR and western blot. (a) The number of TRAP-positive osteoclasts. (b) The expressions of DANCR, Jagged1 and RANKL in PDL tissues. *P < 0.05 vs. si-control.
4 Discussions
In the present study, we demonstrated a significant increase in DANCR expression in the areas of root resorption during experimental tooth movement in rats in vivo and in hPDL cells subjected to CF. The obtained results indicate that abnormal expression of DANCR probably be related to the occurrence of OIIRR. This is the first study on the role of lncRNA DANCR in OIIRR.
LncRNAs have been reported to regulate diverse biological processes, including cell growth, differentiation, chromatin modification, and others [Citation20]. Recently, lncRNAs have been reported to regulate bone formation and osteogenic differentiation of PDL stem cells. He et al. [Citation21] revealed that lncRNA TUG1 promoted osteogenic differentiation of PDL stem cells, and the mechanism might be related to Lin28A. Peng et al. [Citation22] suggested that lncRNA ANCR inhibited bone formation of PDL stem cells through miR-758/Notch2. Moreover, lncRNAs are involved in the osteoblast differentiation, e.g. lncRNA H19 could significantly accelerate in vivo and in vitro osteoblast differentiation via Wnt/β-catenin pathway [Citation23]. However, little is known about the relationship between lncRNAs and hPDL cells-mediated osteoclastogenesis.
hPDL cells are the most common cell type found in the human periodontal ligament. It can not only maintain tissue homeostasis, but also participate in the physiological and pathological molecular processes during OIIRR [Citation24]. In addition, hPDL cells has been considered as a mediator of osteoclastogenesis [Citation25,Citation26]. To investigate the role of DANCR in hPDL cells-mediated osteoclastogenesis, we performed the strategy of loss-and gain-of-function, and found that knockdown of DANCR could reduce the promotion effects of the cell-conditioned medium obtained from the hPDL cells subjected to CF on osteoclastogenesis of hOCPs, which suggested that DANCR could affect hPDL cells-mediated osteoclastogenesis. Meanwhile, the mechanism of DANCR in the process might be related to Jagged1.
Jagged1 is an important protein in Notch signaling pathway, which is located at cytogenetic location 20p12.2 [Citation27]. It has been reported that Jagged1 is involved in the bone development. Lawal et al. [Citation28] demonstrated a critical role of Jagged1 in bone homeostasis, where maintained the transition of osteoprogenitor to maturing osteoblasts. Sekine et al. [Citation29] suggested that inhibition of Jagged1 could suppress osteoclastogenesis via Notch1. Moreover, Manokawinchoke et al. [Citation30] showed that culturing RAW264.7 cells with conditioned medium from Jagged1-treated hPDL cells enhanced osteoclast formation, which might imply a role of Jagged1 on the regulation of osteoclast differentiation via hPDL cells. In the present study, we confirmed the effects of Jagged1 on osteoclastogenesis, and revealed that knockdown of DANCR could inhibit osteoclast formation and bone resorption induced by compression force via down-regulating Jagged1 from hPDL cells, and the mechanism has been confirmed in vivo experiments. Besides that, we found a target relationship between miR-34a-5p and DANCR, and CF could regulate Jagged1 expression in hPDL cells through miR-34a-5p/DANCR.
MiR-34a-5p has been reported to play an important role in the inhibition of osteoclast formation, and it can inhibit osteoclastogenesis and bone resorption by down-regulating Tgif2 [Citation31]. However, our study did not thoroughly explore and verify the role and mechanism of miR-34a-5p in osteoclast formation. Further studies are needed to investigate these using in vitro and in vivo experiments.
In summary, we demonstrated that DANCR response to excessive CF might stimulate the process of osteoclast formation and root resorption via Jagged1 production from hPDL cells. Our results indicate for the first time that DANCR plays an important role in OIIRR, which may provide a new target for reducing OIIRR.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Sawhney R, Sharma R, Sharma K. Microbial colonization on elastomeric ligatures during orthodontic therapeutics: an overview. Turk J Orthod. 2018;31:21–25.
- Asano M, Yamaguchi M, Nakajima R, et al. IL-8 and MCP-1 induced by excessive orthodontic force mediates odontoclastogenesis in periodontal tissues. Oral Dis. 2011;17:489–498.
- Kikuta J, Yamaguchi M, Shimizu M, et al. Notch signaling induces root resorption via RANKL and IL-6 from hPDL cells. J Dent Res. 2015;94:140–147.
- Ashley JW, Ahn J, Hankenson KD. Notch signaling promotes osteoclast maturation and resorptive activity. J Cell Biochem. 2015;116:2598–2609.
- Lindsell CE, Shawber CJ, Boulter J, et al. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–917.
- Nakao A, Kajiya H, Fukushima H, et al. PTHrP induces Notch signaling in periodontal ligament cells. J Dent Res. 2009;88:551–556.
- Xu K, Xu Q, Wu Z. LncRNA NEAT1 is involved in temozolomide resistance by regulating MGMT in glioblastoma multiforme. Clin Surg Res Commun. 2018;2:24–30.
- Wang Y, Luo TB, Liu L, et al. LncRNA LINC00311 promotes the proliferation and differentiation of osteoclasts in osteoporotic rats through the Notch signaling pathway by targeting DLL3. Cell Physiol Biochem. 2018;47:2291–2306.
- Liu C, Cao Z, Bai Y, et al. LncRNA AK077216 promotes RANKL-induced osteoclastogenesis and bone resorption via NFATc1 by inhibition of NIP45. J Cell Physiol. 2018. [Epub ahead of print]. DOI:10.1002/jcp.27031
- Jia J, Li F, Tang XS, et al. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7:37868–37881.
- Ma Y, Zhou G, Li M, et al. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-kappaB signaling pathway. Neurochem Int. 2018;118:233–241.
- Mao Z, Li H, Du B, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37:BSR20171070.
- Zhang J, Tao Z, Wang Y. Long noncoding RNA DANCR regulates the proliferation and osteogenic differentiation of human bone-derived marrow mesenchymal stem cells via the p38 MAPK pathway. Int J Mol Med. 2018;41:213–219.
- Tang Z, Gong Z, Sun X. LncRNA DANCR involved osteolysis after total hip arthroplasty by regulating FOXO1 expression to inhibit osteoblast differentiation. J Biomed Sci. 2018;25:4.
- Irwandi RA, Vacharaksa A. The role of microRNA in periodontal tissue: A review of the literature. Arch Oral Biol. 2016;72:66–74.
- Yu W, Zheng Y, Yang Z, et al. N-AC-l-Leu-PEI-mediated miR-34a delivery improves osteogenic differentiation under orthodontic force. Oncotarget. 2017;8:110460–110473.
- Nakajima R, Yamaguchi M, Kojima T, et al. Effects of compression force on fibroblast growth factor-2 and receptor activator of nuclear factor kappa B ligand production by periodontal ligament cells in vitro. J Periodontal Res. 2008;43:168–173.
- Yamaguchi M, Aihara N, Kojima T, et al. RANKL increase in compressed periodontal ligament cells from root resorption. J Dent Res. 2006;85:751–756.
- Minato Y, Yamaguchi M, Shimizu M, et al. Effect of caspases and RANKL induced by heavy force in orthodontic root resorption. Korean J Orthod. 2018;48:253–261.
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669.
- He Q, Yang S, Gu X, et al. Long noncoding RNA TUG1 facilitates osteogenic differentiation of periodontal ligament stem cells via interacting with Lin28A. Cell Death Dis. 2018;9:455.
- Peng W, Deng W, Zhang J, et al. Long noncoding RNA ANCR suppresses bone formation of periodontal ligament stem cells via sponging miRNA-758. Biochem Biophys Res Commun. 2018;503:815–821.
- Liang WC, Fu WM, Wang YB, et al. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep. 2016;6:20121.
- Kirschneck C, Meier M, Bauer K, et al. Meloxicam medication reduces orthodontically induced dental root resorption and tooth movement velocity: a combined in vivo and in vitro study of dental-periodontal cells and tissue. Cell Tissue Res. 2017;368:61–78.
- Yucel-Lindberg T, Bage T. Inflammatory mediators in the pathogenesis of periodontitis. Expert Rev Mol Med. 2013;15:e7.
- Meikle MC. The tissue, cellular, and molecular regulation of orthodontic tooth movement: 100 years after carl sandstedt. Eur J Orthod. 2006;28:221–240.
- Oda T, Elkahloun AG, Pike BL, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242.
- Lawal RA, Zhou X, Batey K, et al. The notch ligand jagged1 regulates the osteoblastic lineage by maintaining the osteoprogenitor pool. J Bone Miner Res. 2017;32:1320–1331.
- Sekine C, Koyanagi A, Koyama N, et al. Differential regulation of osteoclastogenesis by Notch2/Delta-like 1 and Notch1/Jagged1 axes. Arthritis Res Ther. 2012;14:R45.
- Manokawinchoke J, Sumrejkanchanakij P, Subbalekha K, et al. Jagged1 inhibits osteoprotegerin expression by human periodontal ligament cells. J Periodontal Res. 2016;51:789–799.
- Krzeszinski JY, Wei W, Huynh H, et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512:431–435.