ABSTRACT
Efforts to search for better treatment options for cancer have been a priority, and due to these efforts, new alternative therapies have emerged. For instance, clinically relevant tumor-suppressive microRNAs that target key oncogenic drivers have been identified as potential anti-cancer therapeutics. MicroRNAs are small non-coding RNAs that negatively regulate gene expression at the posttranscriptional level. Aberrant microRNA expression, through misexpression of microRNA target genes, can have profound cellular effects leading to a variety of diseases, including cancer. While altered microRNA expression contributes to a cancerous state, restoration of microRNA expression has therapeutic benefits. For example, ectopic expression of microRNA-34a (miR-34a), a tumor suppressor gene that is a direct transcriptional target of p53 and thus is reduced in p53 mutant tumors, has clear effects on cell proliferation and survival in murine models of cancer. MicroRNA replacement therapies have recently been tested in combination with other agents, including other microRNAs, to simultaneously target multiple pathways to improve the therapeutic response. Thus, we reasoned that other microRNA combinations could collaborate to further improve treatment. To test this hypothesis miR-34a was used in an unbiased cell-based approach to identify combinatorial microRNA pairs with enhanced efficacy over miR-34a alone. This approach identified a subset of microRNAs that was able to enhance the miR-34a antiproliferative activity. These microRNA combinatorial therapeutics could offer superior tumor-suppressive abilities to suppress oncogenic properties compared to a monotherapeutic approach. Collectively these studies aim to address an unmet need of identifying, characterizing, and therapeutically targeting microRNAs for the treatment of cancer.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that negatively regulate gene expression at the posttranscriptional level [Citation1–Citation3]. Aberrant expression of miRNAs is a hallmark of cancer that results in misexpression of miRNA target genes generating profound cellular effects leading to a variety of diseases, including cancer. For example, the tumor-suppressive miRNAs, miRNA-34a (miR-34a), miR-34b, and miR-34c, that collectively belong to the miR-34 family are downregulated in a variety of cancers including non-small cell lung cancer (NSCLC) [Citation4,Citation5]. Downregulation of miRNAs occurs via a variety of mechanisms including genetic alterations [Citation6], aberrant oncogene- and tumor suppressor-mediated transcription [Citation7,Citation8], epigenetic mechanisms [Citation9,Citation10] and defects in miRNA biogenesis [Citation11–Citation13]. For instance, in both NSCLC and breast cancer, the alleles encoding the three miR-34 family members are deleted due to their location in fragile regions of the genome [Citation14]. The three miR-34 family members are also subject to epigenetic inactivation by CpG methylation [Citation9,Citation15]. Clinically, promoter hypermethylation of miR-34a was found to be a poor predictor of NSCLC patient relapse [Citation5]. Reduced levels of miR-34a, miR-34b, and miR-34c also occur due to mutations in p53, a tumor-suppressive transcription factor that directly regulates the three family members [Citation7,Citation16–Citation19]. Thus, in p53 mutant tumors, miR-34 levels are markedly reduced. In addition to miR-34, various other miRNAs are depleted in human carcinogenesis, many of which have tumor-suppressive function. Through concerted efforts, many of these tumor suppressive miRNAs have been tested as anti-cancer therapeutics with some promising results [Citation20,Citation21].
MiRNA replacement therapeutics aim to restore the expression of a miRNA that has been lost or downregulated in a cell, with multiple studies reporting a therapeutic benefit [Citation22–Citation24]. For instance, reintroducing miR-34a in various murine models of cancer has clear effects on reducing cell proliferation and increasing survival [Citation20–Citation22,Citation25,Citation26]. While there are many mechanisms used to reintroduce miRNAs in vivo [Citation27], one clinically feasible approach involves the use of miRNA synthetic analogs that mimic mature miRNAs (so-called mimics) obviating the need to use the endogenous miRNA biogenesis machinery. Indeed, miR-34a was the first miRNA mimic to enter into clinical trial in early 2013 [Citation28,Citation29] and although the trial was met with some unforeseen severe immune-related adverse events, efforts are still underway to advance miRNAs clinically including their use as sensitizers to other standard-of-care agents [Citation30].
MiRNA replacement therapies have recently been tested in combination with other agents, small molecules and biologics, to simultaneously target multiple pathways to improve the therapeutic response [Citation30–Citation34]. In addition, miRNAs have also been tested in combination with other miRNAs [Citation22,Citation26]. Since miRNAs can regulate multiple gene targets [Citation35,Citation36] it is possible that two or more miRNAs could collaborate to repress the expression of numerous cancer-related genes in various pathways. Our lab and others have shown that it is possible to combine two small RNAs to enhance the therapeutic response [Citation22,Citation26]. For instance, we showed that miR-34a can cooperate with let-7b, another tumor-suppressive miRNA that targets Kras, in KrasG12D/+;p53−/- murine non-small cell lung cancer (NSCLC) [Citation22]. Thus, we reasoned that additional miRNAs could potentially collaborate with miR-34a to repress the expression of other relevant cancer-related genes affecting multiple pathways at once, and thus further improving therapeutic responses.
In this study, miR-34a was evaluated in the presence of 2,019 individually arrayed human-encoded miRNAs for miRNAs that enhanced the miR-34a effect in NSCLC cells. miR-34a was selected as the candidate miRNA for multiple reasons. Firstly, miR-34a is located in a fragile region of the genome often deleted in NSCLC [Citation14]. Because of this, NSCLC patients often present with low levels of miR-34a. Secondarily, all of the miR-34 family members are transcriptionally regulated by p53 [Citation7,Citation16–Citation19], arguably the most mutated gene in NSCLC [Citation37] and globally in human carcinoma [Citation38,Citation39]. Thus, identification of miR-34a-synergistic miRNAs would not only inform future NSCLC therapeutic potential but also therapeutics for other p53-mutated cancers. Thirdly, our previous data in NSCLC mouse models highlight a striking therapeutic effect upon miR-34a restoration at preventing NSCLC and reducing its progression, whether miR-34a is delivered directly to the lung in lentiviral particles [Citation23], or systematically by way of lipid vesicles [Citation22] or ligand conjugates [Citation25,Citation40]. Fourthly, miR-34a was the first miRNA-restoration therapy to enter into clinical trial [Citation29]. While the trial was ultimately not successful, it is anticipated that future miRNA mimic-based trials will include miR-34a due to its strong tumor-suppressive activity in various tumor subtypes.
Herein, we used an unbiased cell-based approach to identify combinatorial miRNA pairs with enhanced efficacy over miR-34a alone. This approach allowed us to identify a subset of miRNAs that was able to enhance miR-34a antiproliferative activity in NSCLC cell lines. Our evaluation of the top candidate pairs identified synergistic interactions of miR-34a with five miRNAs. KEGG analysis and target predictions for these five miRNAs predicted multiple cancer-related pathways that the miRNAs function in and a few oncogenic targets that may to be shared between a subset of the miR-34a-synergistic miRNAs. These miRNA combinatorial therapeutics could offer superior tumor-suppressive abilities to suppress oncogenic properties compared to a monotherapeutic approach. We propose that by targeting multiple oncogenic drivers at the same time using miRNA combinations that act synergistically, the development of drug resistance is less likely due to the inability to accumulate mutations in the various targets and due to a reduced number of cancer-related bypass tracks available for the cancer cell to engage.
Materials and methods
Cell culture
A panel of six non-small cell lung cancer (NSCLC) cell lines: H441, A549, EKVX, Calu6, H460, and H23, mycoplasma-free as determined by testing for mycoplasma contamination via MycoAlert Mycoplasma Detection Kit (Lonza), were grown in RPMI 1640 medium (Life Technologies) supplemented with 10% fetal bovine serum (Sigma), penicillin (100 U/ml), and streptomycin (100 g/ml) (HyClone, GE Healthcare Life Sciences), and maintained at 37°C in 5% CO2. Authentication of cell lines was performed using short tandem repeat profiling [American Type Culture Collection (ATCC)].
H441-pmiR cell line generation for measuring transfection efficiency
H441 cells were transfected with pmiR-Glo plasmid (Promega) and stable clones (H441 pmiR) were isolated after G418 antibiotic selection. Stable clones were grown in RPMI 1640 medium supplemented with G418 (300 ug/mL), 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 g/ml), and maintained at 37°C in 5% CO2. H441-pmiR clones were screened for luciferase activity using the Dual Glo Luciferase Assay (Promega) following the instructions of the manufacturer (Figure S1). Luciferase signal was measured in a GloMax plate reader (Promega). Firefly luciferase is used as a measurement of the transfection efficiency when cells are co-transfected with a siRNA against firefly luciferase (siLuc2) and it is calculated as a percentage of firefly activity in untreated cells. Wells with <90% knockdown of firefly luciferase were excluded from analyses. Renilla activity serves as a proxy for cell number ()).
miR-34a dose response studies
Transfections with miR-34a were performed to determine the dose of mimic to be used in the screen. Transfections were performed in 384-well plates using a reverse transfection protocol with Lipofectamine RNAimax (Invitrogen) using the following concentrations: 0.5, 1.5, 3, 6, 12 and 25 nM. A scrambled RNA was used as a negative control (PremiR-NC2, Ambion). Each transfection was supplemented with negative control such that all transfections were performed with a final concentration of 25 nM. Briefly, miRNA mimics were diluted in serum-free medium (SFM) to a final volume of 5 uL (per well). In a separate tube, 0.05 uL of Lipofectamine RNAimax were added to SFM to a final volume of 5 uL (per well). Next, 5 uL of the miRNA mix and 5 uL of Lipofectamine RNAimax solution were mixed and incubated at room temperature for 30 min. H441 cells were harvested and cells were resuspended at 4 × 104 cells/mL. Ten microliters of the miRNA/Lipid cocktail were added to the bottom of the wells of a 384-well plate (six wells per treatment). Next, 10 uL of the cell suspension was added to each well (20 uL/well final total volume). Plates were incubated at 37°C with 5% CO2 for five days. On day five, cell proliferation was evaluated using luciferase signal as a proxy for cell number as described previously.
miRNA library screening
The human mirVana miRNA Library (Invitrogen; based on miRBase v. 21) was used to evaluate 2,019 human-encoded miRNAs in a cell-based screen. First, each individual miRNA was assayed in combination with miR-34a at equimolar concentration (3nM each, total RNA 6 nM). Transfections were performed as previously described in 384-well plates using the reverse transfection protocol with Lipofectamine RNAimax (Invitrogen) [Citation41]. The following controls were included in each plate: i) miRNA being evaluated + miR-34a (3 nM each), ii) full dose of miR-34a (6 nM), iii) the full dose of a scrambled miRNA control (6 nM), and iv) untreated. On day five, cell proliferation was evaluated using the luciferase signal as a proxy for cell number as described previously. Outliers were eliminated if transfection efficiency was determined to be lower than 90%. Cell proliferation was normalized to negative control (PremiR-NC2) transfected cells and miRNA combinations were ranked according to the normalized values. To identify putative positive hits, ± three standard deviations of the full dose of mir-34a (6 nM) was used as the threshold.
Secondary validation of putative hits
A secondary validation of putative hits identified from the primary screening was performed using the sulforhodamine B (SRB) assay as a proxy for cell number [Citation41]. The following controls were included in each plate: miRNA being evaluated + miR-34a (3 nM each), ii) full dose of miR-34a (6 nM), iii) the full dose of a scrambled miRNA control (6 nM, PremiR-NC2, Ambion) and iv) the full dose of each miRNA being evaluated (6 nM). Each of the transfections was performed using the following panel of non-small cell lung cancer (NSCLC) cell lines: H441, A549, EKVX, Calu6, H460 and H23. The Response Additivity approach [Citation42] was used to determine which miRNA combinations were better than each miRNA alone. The approach consists of showing that the effect of a positive miRNA combination occurs when the combination effect (Epair) is greater than the expected additive effect given by the sum of the individual effects (Combinatorial index (CI) = (EmiR-34a + EmiR-X)/Epair) [Citation43]. The statistical significance is then given by the p-value (p = 0.05) of the Student’s T-test comparing the miRNA combination effect to the effect of miR-34a as a single agent. Since this approach provides limited evidence of synergy, additional experiments were performed to evaluate cell proliferation of H441 cells transfected with different concentrations of the miRNA combination and the individual miRNAs. Transfections with miRNA candidates were performed in 384-well plates at a ratio of 1:1 (miR-34a + miR-X) using the following concentrations: 0.5, 0.75, 1.5, and 3 nM. Each transfection was supplemented with negative control to a final concentration of 6 nM. Synergism was evaluated using Combosyn software (Combosyn) based on the combination index (CI)-isobologram equation [Citation44]. The equation helps to gain a better understanding of drug interactions, where CI < 1 indicate synergism, CI = 1 indicate an additive effect, and CI > 1 indicate antagonism [Citation44].
Bioinformatic analysis of miR-34a-synergistic miRNAs
Predicted targets for the five miR-34a-synergistic miRNAs were obtained using TargetScan [Citation45] (miR-4664-3p, miR-5100) or an overlap of targets shared between TargetScan and miRDB [Citation46] (miR6715-5p, miR-3157-3p and let-7a-2-3p). Each of the five miRNAs were also evaluated for pathway enrichment using miRNA Pathway Dictionary Database (miRPath DB v1.1). For both analyses, significantly enriched pathways were graphed using -Log (p-value).
Results
Cell line generation and response to miR-34a
To identify miRNAs that synergize with miR-34a, an unbiased screen of miR-34a in combination with 2,019 individually arrayed human-encoded miRNAs was performed. Because miRNA mimics were transfected into cells, it was essential to develop a suitable assay that would allow for the normalization of transfection variabilities between wells ()). To this end, H441 non-small cell lung cancer cells were generated that expresses both firefly and renilla luciferase (H441-pmiR). Firefly was used to monitor transfection efficiency following transfection of cells with a low concentration of an siRNA targeting firefly (siLuc2). Renilla was used as a surrogate for cell number to monitor the effect the miRNA(s) had on cell growth/death. Multiple H441-pmiR stable clones were isolated (Supplemental Figure 1) and the clone with the highest firefly and renilla levels was selected for the screen. Using this clone, it was determined that both renilla and firefly activity correlate with cell number reaching an R2 of 0.92 and 0.98, respectively ()) and that low doses of siRNA against luciferase (siLuc2) cause a significant decrease in firefly activity ()). Based on this assessment, 0.5 nM of siLuc2 was co-transfected with every miRNA combination and firefly luciferase was used as a measurement of the transfection efficiency measured as a percentage of firefly repression compared to untreated cells. Renilla activity was used as a proxy for cell number ()). Next, it was determined that when the dose of miR-34a transfected into H441 cells exceeded 6 nM there was no further reduction in cell proliferation ()). This dose is in stark contrast to most published studies that often use doses of 50–200 nM to evaluate the effects of miRNA mimics. These high doses are likely causing off-target effects and/or effects that would not be achievable in a cell following a therapeutic dose. Because we foresee future advancement of these combinations into the clinic 6 nM was chosen as the highest dose. The selected clone was further evaluated for firefly and renilla activity in the presence of siLuc2 and/or miR-34a at the predetermined concentrations that would be used in the screen. As expected, co-transfecting siLuc2 reduced firefly activity ()) while transfection of a tumor-suppressive miRNA, miR-34a reduced renilla levels ()), signifying a reduction in total cell number in accordance with the model in ).
Identification of miRNAs that synergize with miR-34a
Using the H441-pmiR clone, miRNAs that enhanced miR-34a activity were identified through a screen. H441-pmiR cells were transfected with miR-34a at 6 nM (full dose) or with miR-34a (3 nM) and every other individual miRNA (3 nM) and changes in renilla activity were determined. Renilla activity was then normalized to firefly activity to account for inter-well variability in transfection. Data from all 2,019 miRNAs in combination with miR-34a are arrayed from least (left) to most (right) effective ()). The effect of miR-34a combined with itself (6 nM) is plotted with three standard deviations above and below the miR-34a effect indicated. To verify that the screen worked as intended, ~250 miRNAs from both the right- and left-most portions of the graph were identified in the literature as oncogenic, tumor suppressive, or inconclusive, color-coded green, red, or orange, respectively, in ). Some of the well-reported oncogenic miRNAs, such as miR-9-5p, miR-21 and miR-155 were clustered on the left side of the graph suggesting that they may be antagonizing miR-34a function, while tumor-suppressive miRNAs, such as let-7c, miR-98, and miR-29c localized on the right side of the graph clustering with miRNAs that reduced cell proliferation. There was a clear enrichment of previously reported tumor-suppressive miRNAs clustering on the right side of the graph (80% of the miRNAs with reported function) relative to only 29% of previously reported tumor-suppressive genes being identified on the left, again supporting the validity of the screen.
The results of the primary screen indicate that there is a subset of miRNAs that when combined with miR-34a can reduce cell proliferation levels compared to cells transfected with mir-34a alone (6 nM, )). Since the 6 nM dose of each miRNA from the library was not included in the primary screen there was a possibility that the effect of the combination could be due to the miRNA alone, and not due to its combined activity with miR-34a. Thus, the miRNA combinations that fell below 3 standard deviations of the effect achieved by miR-34a alone (6 nM, 333 combinations, blue-shaded box )) were re-evaluated. For the validation assay, each miRNA was again tested with miR-34a (3nM each) and the miRNA candidates were evaluated alone (6 nM). A miRNA that worked cooperatively with miR-34a would have a better antiproliferative effect when combined with miR-34a than the miRNA being tested on its own (111 miRNA pairs, blue box )). To provide further evidence of the superiority of the miRNA combination relative to the individual miRNA an additional functional assay was used to test the 111 miRNA pairs in a panel of six NSCLC cell lines (H441, A549, EKVX, Calu6, H460, H23). An SRB assay took the place of the luciferase assay to evaluate cell proliferation. The assay was validated for linearity with regard to cell density ()) and miR-34a response ()). The Response Additivity approach [Citation42] was used to determine which miRNA combinations were better than each miRNA alone. MiRNA combinations that had a CI lower than 1, p < 0.05 and had normalized proliferation values of >1 when the miRNA was tested alone were shortlisted for each of the cell lines. The lists of miRNAs obtained from each NSCLC cell line was compared and 12 miRNA combinations that enhanced miR-34a effect in at least four different cell lines were identified ()).
Next, the top 12 miRNA combinations were further evaluated by assessing the potencies of the individual miRNA, miR-34a, or the combination and characterizing the possible existence of synergistic or antagonistic effects. The data shows that there are five miRNAs that have higher antiproliferative activity when combined with miR-34a: miR-6715b, miR-4664-3p, miR-3157-3p, miR-5100 and let-7a-2-3p (): labeled, –)). In agreement with this observation, the CI values for these combinations are <1, thus suggesting the possibility of a synergistic interaction ()).
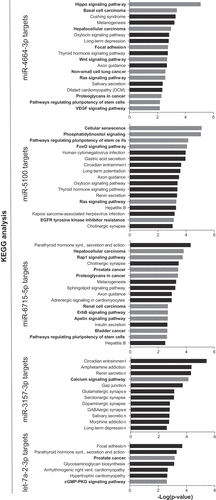
Various bioinformatic analyses were conducted to support the experimental findings. Firstly, using literature-based software analyses (TargetScan [Citation45] alone or TargetScan and miRDB [Citation46]) miRNA targets were predicted for the synergistic miRNAs. The predicted targets were then used for KEGG pathway enrichment analysis to identify cellular pathways that the predicted miRNA targets were enriched in (). For miR-4664-3p and miR-5100, overlapping targets from TargetScan and miRDB did not identify significantly enriched pathways; thus, to gain insight into potential pathways that miR-4664-3p and miR-5100 regulate only TargetScan was used for KEGG analysis. For miR-6715b, miR-3157-3p, and let-7a-2-3p overlapping targets identified in both databases were used for the analysis. miRNA Pathway Dictionary Database (miRPathDB v1.1) [Citation47,Citation48] identified a similar, but a more extensive list of KEGG pathways (Supplemental Figure 2), albeit p-values were markedly lower. Notably, several cancer-related pathways were identified including focal adhesion (cell motility), EGFR tyrosine kinase inhibitor resistance, and proteoglycans in cancer, as well as a number of growth factor signaling pathways such as those involving RAS and Hippo (major cancer-related pathways/functions are depicted as gray bars in ). This implicates that miRNAs that synergize with miR-34a likely regulate multiple facets of cancer cell biology. Indeed, when the targets of the five miRNAs were combined and Ingenuity Pathway Analysis (IPA) was performed the findings were corroborated, the targets overwhelming identified cancer as the top disease modulated with a p-value of 8.88 × 10−31. In line with the KEGG analysis, IPA identified the top molecular functions of the targets as cellular movement (p-value 8.86 × 10−7), gene expression (p-value 6.31 × 10−7), cell death and survival (p-value 8.86 × 10−7) and cell cycle (p-value 8.94 × 10−6).
Secondarily, we sought to identify individual genes that are predicted to be targets of more than one of the five miR-34a-synergistic miRNAs. These genes represent potential critical target genes that may largely contribute to the synergistic anti-proliferative effect. Genes that were predicted to be the targets of at least three miRNAs are defined in . Most of the shared targets have reported functions as oncogenes or are involved in therapeutic resistance, and have been found to be upregulated in human cancers. Because most of the miRNAs identified from the combinatorial screen are presumably tumor-suppressive miRNAs, it is expected that most of their target genes should be promoting cancer cell proliferation and/or resistance.
Table 1. Genes predicted to be the targets of at least three miR-34a synergistic miRNAs.
Discussion
The results of this study indicate that miRNA combinatorial therapeutics could offer superior tumor-suppressive abilities in lung cancer cells compared to a monotherapeutic approach. The combination of each of the top five candidate miRNAs with miR-34a was able to enhance miR-34a antiproliferative activity and further reduce cell proliferation. Our evaluation of these five candidate pairs showed that they have a synergistic interaction, meaning that the combination works better than each miRNA as a single agent. It is likely that these miRNA combinations could be used clinically in scenarios in which drug resistance to monotherapies often develops due to the accumulation of mutations. We propose that targeting multiple oncogenic drivers with miRNAs that act synergistically could be beneficial to cancer therapy avoiding the development of resistance.
Indeed, the predicted targets of the top five miR-34a-synergistic miRNAs are enriched for various pathways that are involved in promoting cancer: RAS, VEGF, WNT, HIPPO, EGFR, ErbB and others. Shortlisting the targets resulted in the identification of two predicted targets regulated by four of the five miRNAs, both of which are genes involved in pro-growth, regulating proliferation and cell-cycle. An additional 13 targets are predicted to be regulated by three of the five miR-34a-synergistic miRNAs, where eight of the 13 have pro-tumorigenic functions. The remaining five did not have any publications on their cellular roles, so an accurate prediction of their function could not be made. In future studies, it will be essential to determine if the miR-34a-synergistic miRNAs are capable of regulating these predicted targets and if downregulation of these specific targets is involved in the synergistic response.
While this study shows that there are miRNAs that could be used in combination with miR-34a to enhance antiproliferative abilities in cell-based studies, further in vivo validation will be required, particularly focused on a comparison between the monotherapies and the combinatorial miRNA therapeutics. One possible model that could be used for these types of studies is the Kras/p53 mouse model that has been used in previous studies to access the therapeutic potential of miR-34a [Citation22,Citation23]. Due to the fact that some of these miRNA candidates are reported to be the passenger strand, a transgene overexpression approach (viral delivery for instance) would not be feasible since it would require processing of the pre-miRNA into the mature form and there is no guarantee that the correct strand would be loaded into the RNA induced silencing complex (RISC). Perhaps an approach using chemically synthesized and modified miRNA mimics with either a lipid- or ligand-based [Citation21,Citation36–Citation38] delivery platform would be useful to ensure delivery and loading of the correct strand into RISC.
It is also important to take notice of the large number of miRNAs that appear to antagonize miR-34a activity ()). This information certainly brings into question the response of tumors to miR-34a therapeutics based on the genetics of the tumor, in particular with regard to the levels of other miRNAs. In the future, a more personalized approach to miR-34a-based therapeutics may be essential to stratify responders from potential non-responders and further argues that a combinatorial approach may increase the number of responders.
Figure 1. Screening approach. (a) Use of a dual luciferase expression vector to evaluate transfection efficiency and cell number. The schematic depicts a model in which firefly luciferase is used as a measurement of the transfection efficiency when cells are co-transfected with a siRNA against firefly luciferase (siLuc2) and a miRNA. Renilla activity serves as a proxy for cell number and its activity is only affected when a miRNA that alters cell proliferation is transfected. (b) A linear response between cell number and Renilla and Firefly luciferase activity (Error bars: mean ± s.d., n = 6). C) Firefly luciferase repression upon transfection with siLuc2. Error bars: mean ± s.d. Each experiment corresponds to n = 6 per treatment. (d) miR-34a dose response in H441-pmiR cells. Error bars: mean ± s.d. Each experiment corresponds to n = 8 per treatment. Statistical analysis performed with two-way ANOVA with post hoc Bonferroni correction (***, P < 0.001). (e) Firefly and (f) Renilla activity following transfection with miR-34a or PremiR-NC2 in presence of siLuc2. Error bars: mean ± s.d. Each experiment corresponds to n = 6 per treatment. Statistical analysis for B, C, D, E and F performed with a one-way ANOVA with post hoc Bonferroni correction (*, P < 0.05; **, P < 0.01; ****, P < 0.0001). RLU: relative light units.
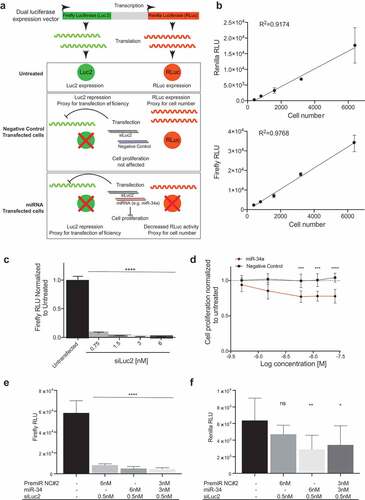
Figure 2. miRNAs alter miR-34a antiproliferative potential. (a) miRNA mimics ranked according to effect, normalized to negative control (NC) transfected cells. Red dashed line = 3 s.d. of the effect of miR-34a alone. Black dashed line = effect of NC treated cells. Blue shaded box = 333 miRNA mimics that were moved forward in B. n = 6. Each dot represents the mean effect of a miRNA combination. (b) Validation of 333 miRNA mimics represented as the effect of the individual miRNA (Esingle) minus the effect when combined with miR-34a (Epair). Values <0 indicate that the combination has a better antiproliferative activity than the miRNA by itself. n = 6. Each dot represents the mean effect of a miRNA combination. Blue shaded box = 111 miRNA mimics with apparent combinatorial activity. The five miRNAs subsequently determined to have synergistic activity with miR-34a are indicated.
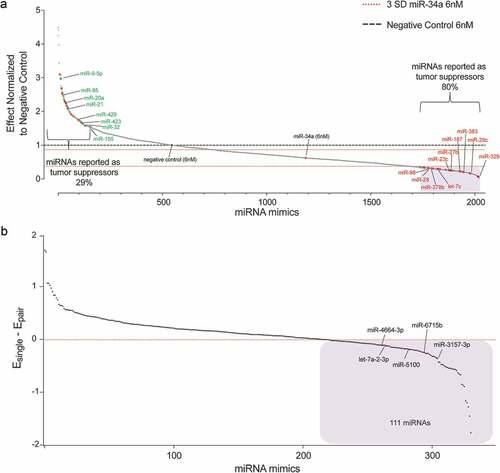
Figure 3. Validation of a combinatorial effect of the 111 miRNA mimics in combination with miR-34a in a panel of NSCLC cell lines. (a) Validation of sulforhodamine B (SRB) method for secondary verification of screen hits. Absorbance of SRB as a linear response to H441-pmiR cell number. Error bars: mean ± s.d., n = 6. (b) Response of H441-pmiR to miR-34a measured by SRB. Error bars: mean ± s.d., n = 6. (c) miRNA combinations evaluated in a panel of six NSCLC cell lines. miRNAs included in this comparison had a CI lower than 1 (potential synergism), p < 0.05 and cell proliferation of the single miRNA was <1.
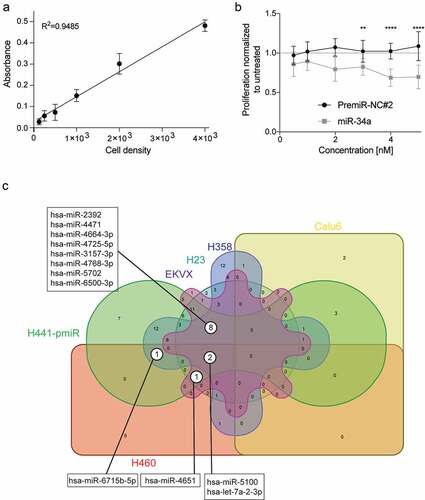
Figure 4. Evaluation of synergism of positive hits. The possibility of synergism with miR-34a was evaluated for (a) miR-6715b, (b) miR-4664-3p, (c) miR-3157-3p, (d) miR-5100 and (e) let-7a-2-3p. (For A-E. red lines: miR-34a alone, blue lines: putative synergistic miRNA alone, green lines: combination) miRNA combinations have a stronger effect than each miRNA on its own. (f) Evaluation of synergism using the (CI)-isobologram equation. The equation helps to gain a better understanding of drug interactions, where CI < 1 indicate synergism. Statistical analysis performed with one-way ANOVA with post hoc Bonferroni correction (***, P < 0.001; ****, P < 0.0001).
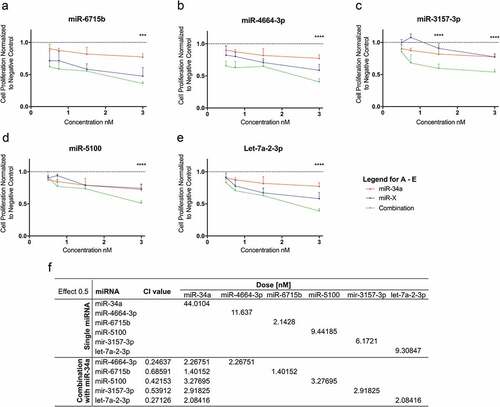
Figure 5. Pathway enrichment analysis of miR-34a-synergistic miRNA target genes. Predicted targets for the five synergistic miRNAs were determined and used to identify pathways that the target genes are enriched in. Cancer-related pathways/diseases are in bold text with corresponding bars depicted in grey.
Supplemental Material
Download MS Word (1.9 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862.
- Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906.
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854.
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199.
- Gallardo E, Navarro A, Viñolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909.
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866.
- Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307.
- O’Donnell KA, Wentzel EA, Zeller KI, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843.
- Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600.
- Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443.
- Melo SA, Moutinho C, Ropero S, et al. A genetic defect in Exportin-5 traps precursor MicroRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315.
- Hill DA, Ivanovich J, Priest JR, et al. DICER1 mutations in Familial pleuropulmonary blastoma. Science. 2009;325:965.
- Piskounova E, Polytarchou C, Thornton JE, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079.
- Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004.
- Toyota M, Suzuki H, Sasaki Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132.
- Chang T-C, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752.
- He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–11101.
- Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743.
- Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593.
- Kim M, Kasinski AL, Slack FJ. MicroRNA therapeutics in preclinical cancer models. Lancet Oncol. 2011;12:319–321.
- Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864.
- Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2015;34:3547–3555.
- Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72:5576–5587.
- Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–1122.
- Orellana EA, Tenneti S, Rangasamy L, et al. FolamiRs: ligand-targeted, vehicle-free delivery of microRNAs for the treatment of cancer. Sci Transl Med. 2017;9:eaam9327.
- Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer. Proc Nat Acad Sci. 2014;111:E3553–61.
- Myoung SS, Kasinski AL. CHAPTER 14: strategies for safe and targeted delivery of microRNA therapeutics. In: microRNAs in diseases and disorders. Royal Society of Chemistry 2019. p. 386–415.
- Bader AG. miR-34 – a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120.
- Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577.
- Stahlhut C, Cycle FSC. Combinatorial action of microRNAs let-7 and miR-34 effectively synergizes with erlotinib to suppress non-small cell lung cancer cell proliferation. Cell Cycle. 2015;14:2171–2180.
- Zhang L, Yang X, Lv Y, et al. Cytosolic co-delivery of miRNA-34a and docetaxel with core-shell nanocarriers via caveolae-mediated pathway for the treatment of metastatic breast cancer. Sci Rep. 2017;7:46186.
- Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013;4:e659.
- Deng X, Cao M, Zhang J, et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35:4333–4344.
- Shi S, Han L, Deng L, et al. Dual drugs (microRNA-34a and paclitaxel)-loaded functional solid lipid nanoparticles for synergistic cancer cell suppression. J Control Release. 2014;194:228–237.
- Lewis BP, Shih I-H, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798.
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672.
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380.
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nature Reviews Cancer. 2009;9:749-758.
- Nigro JM, Baker SJ, Preisinger AC, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705–708.
- Orellana EA, Abdelaal AM, Rangasamy L, et al. Enhancing microRNA activity through increased endosomal release mediated by nigericin. Mol Ther Nucleic Acids. 2019;16:505–518.
- Orellana E, Kasinski A. Sulforhodamine B (SRB) assay in cell culture to investigate cell proliferation. Bio-Protocol. 2016;6(21).
- Slinker BK. The statistics of synergism. J Mol Cell Cardiol. 1998;30:723–731.
- Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3:e00149.
- Chou T-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681.
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:101.
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–52.
- Kehl T, Backes C, Kern F, et al. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget. 2017;8:107167–107175.
- Backes C, Kehl T, Stöckel D, et al. miRPathDB: a new dictionary on microRNAs and target pathways. Nucleic Acids Res. 2017;45:D90–6.
- Porayette P, Gallego MJ, Kaltcheva MM, et al. Differential processing of amyloid-beta precursor protein directs human embryonic stem cell proliferation and differentiation into neuronal precursor cells. J Biol Chem. 2009;284:23806–23817.
- Kloet DEA, Polderman PE, Eijkelenboom A, et al. FOXO target gene CTDSP2 regulates cell cycle progression through Ras and p21(Cip1/Waf1). Biochem J. 2015;469:289–298.
- Winans S, Flynn A, Malhotra S, et al. Integration of ALV into CTDSPL and CTDSPL2 genes in B-cell lymphomas promotes cell immortalization, migration and survival. Oncotarget. 2017;8:57302–57315.
- Mittal S, Aslam A, Doidge R, et al. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-not complex contribute to the prevention of cell death and senescence. Mol Biol Cell. 2011;22:748–758.
- Demidyuk IV, Shubin AV, Gasanov EV, et al. Alterations in gene expression of proprotein convertases in human lung cancer have a limited number of scenarios. PLoS One. 2013;8:e55752.
- Deb S, Xu H, Tuynman J, et al. RAD21 cohesin overexpression is a prognostic and predictive marker exacerbating poor prognosis in KRAS mutant colorectal carcinomas. Br J Cancer. 2014;110:1606–1613.
- Supernat A, Lapińska-Szumczyk S, Sawicki S, et al. Deregulation of RAD21 and RUNX1 expression in endometrial cancer. Oncol Lett. 2012;4:727–732.
- Atienza JM, Roth RB, Rosette C, et al. Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol Cancer Ther. 2005;4:361–368.
- Pinho AV, Van Bulck M, Chantrill L, et al. ROBO2 is a stroma suppressor gene in the pancreas and acts via TGF-β signalling. Nat Commun. 2018;9:5083.
- Hsu T-I, Wang M-C, Chen S-Y, et al. Sp1 expression regulates lung tumor progression. Oncogene. 2012;31:3973–3988.
- Beishline K, Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. Febs J. 2015;282:224–258.
- Goffin V. Prolactin receptor targeting in breast and prostate cancers: new insights into an old challenge. Pharmacol Ther. 2017;179:111–126.
- Wang S-M, Tie J, Wang W-L, et al. POU2F2-oriented network promotes human gastric cancer metastasis. Gut. 2016;65:1427–1438.
- Hodson DJ, Shaffer AL, Xiao W, et al. Regulation of normal B-cell differentiation and malignant B-cell survival by OCT2. Proc Nat Acad Sci. 2016;113:E2039–46.
- Du P, Wang S, Tang X, et al. Reduced expression of Metastasis Suppressor-1 (MTSS1) accelerates progression of human bladder uroepithelium cell carcinoma. Anticancer Res. 2017;37:4499–4505.
- Xie F, Ye L, Ta M, et al. MTSS1: a multifunctional protein and its role in cancer invasion and metastasis. Front Biosci Schol. 2011;3:S:621–631.
