ABSTRACT
Background: Hepatocellular carcinoma (HCC) afflicts more than half a million people each year worldwide. It was reported that circ_0015756 was up-regulated in HCC, but the mechanism did not extensively studied.
Methods: we collected 24 paired cancerous and noncancerous liver tissues surgically resected from HCC patients. HCC cell proliferation, invasion, migration and apoptosis in vitro were evaluated using MTT assay, Transwell assay, scratch test and Annexin-V/PI staining respectively. Interactions between circ_0015756 and miR-7, miR-7 and FAK were further validated by the luciferase reporter assay. Tumor xenografts of HCC cells with circ_0015756 knockdown were established in nude mice.
Results: The expression level of circ_0015756 was increased and the expression level of miR-7 was diminished in cancerous liver tissues relative to noncancerous liver tissues. Circ_0015756 knockdown was shown to increase the expression of miR-7, reduce the proliferation, invasion, migration and resistance to apoptosis, and down-regulate the expression of FAK in HCC. We found miR-7 impaired expression of FAK to inhibit HCC cells, suggesting that miR-7 is responsible for the dysfunction of FAK. Importantly, we showed circ_0015756 could up-regulate FAK via targeting miR-7. These in vitro findings were reproduced in vivo that circ_0015756 knockdown decreased HCC xenograft growth.
Conclusion: Our present study reveals a model of HCC development that is composed of circ_0015756, miR-7 and FAK. Modulation of their levels exhibits a promise in the treatment of HCC.
Abbreviations: HCC: hepatocellular carcinoma; circRNAs: circular RNAs; miRNA/miR: microRNA; miR-7: microRNA-7; FAK: focal adhesion kinase; KLF-4: kruppel like factor 4; DKK1: dickkopf WNT signaling pathway inhibitor 1; ccRCC: clear cell renal cell carcinoma; PI3K: phosphoinositide 3-kinase; Ct: comparative threshold cycle; RPMI: Roswell Park Memorial Institute; FBS: fetal bovine serum; RT: reverse transcription; qPCR: quantitative polymerase chain reaction; RIPA: radioimmunoprecipitation assay; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; PVDF: polyvinylidene difluoride; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; MTT: 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; DMSO: dimethyl sulfoxide; DMEM: Dulbecco’s modified Eagle’s medium; PI: propidium iodide; SPF: specific pathogen-free; SD: standard deviation; p-Akt: phosphorylated-Akt; shRNAs: small hairpin RNAs; 3’UTR: 3’-untranslated regions
Introduction
Although the incidence and mortality of liver cancer have been decreasing in China, population growth and aging still cause a heavy burden for a large increasing number of newly diagnosed cases in 2015 [Citation1]. Hepatocellular carcinoma (HCC) is the major subtype of primary liver cancers, accompanied by a poor prognosis [Citation2]. New molecular-targeted therapeutic strategies are urgently needed for improving the treatment outcomes of HCC through identification of biomarkers or targets via investigations associated with molecular cell biology [Citation3,Citation4]. Meanwhile, complex genomic and epigenetic alterations are implicated in the pathogenesis and development of HCC, which poses challenges and obstacles in the molecular classifications [Citation5]. Recent studies have identified potential correlation of a family of endogenous noncoding RNAs, circular RNAs (circRNAs) with the pathogenesis of various human cancers [Citation6,Citation7]. Strikingly, circRNAs exert effects in cellular biological activities, commonly working as microRNA (miRNA or miR) to sponge corresponding direct target genes and result in reduced translation [Citation8]. A newly identified circRNA, circ_0015756, has been suggested to be highly expressed in hepatoblastoma [Citation9]. However, the molecular mechanism underlying the role of circ_0015756 in HCC remains to be investigated.
Functionally, miRNAs are dysregulated in multiple human malignancies, acting as anti-oncomiRs or oncomiRs [Citation10]. It is interesting to note that miR-7 functions as a tumor suppressor in human cancers, such as pancreatic carcinoma [Citation11] and non-small cell lung cancer [Citation12]. More importantly, evidence has demonstrating the potent tumor suppressive role of miR-7 in human HCC [Citation13]. CircRNAs have the ability to act as modulators of miRNA activity in cancers [Citation14]. For example, a novel circRNA Cdr1as was revealed to promote HCC progression through targeting miR-7 [Citation15]. Another circRNA ciRS-7 was demonstrated to induce hepatic microvascular invasion partly via acting as a sponge of miR-7 [Citation16]. Interestingly, miR-7 could hinder the pathological process of HCC by inducing kruppel like factor 4 (KLF-4) mRNA degradation [Citation17].
Notably, this current study identified the putative binding sites between miR-7 and focal adhesion kinase (FAK). A prior study has proved that miR-7 could curtail the local invasion and metastatic potential of breast cancer through targeting and negatively regulating FAK [Citation18]. Recent evidence has also documented the high expression level of FAK in HCC, which was revealed to share associations with tumor progression and metastasis in HCC [Citation19]. Furthermore, FAK was highlighted to interact with activated Akt to accelerate cell migration and invasion in liver cancer [Citation20]. Akt, also known as protein kinase B, is a downstream kinase of phosphoinositide 3-kinase (PI3K) pathway, frequently hyperactivated in human cancers [Citation21].
On the basis of those findings, we propose a hypothesis that circ_0015756, miR-7, FAK and Akt may involve in the HCC progression. However, how they function in HCC cell activities and how they interact with each other remain to be largely unknown. In this present study, a series of loss- and gain- of function assays have been performed to characterize the effect of circ_0015756 on HCC cells. The knockdown of circ_0015756 acted to inhibit the expression of FAK by up-regulating the tumor suppressor miR-7 through the Akt pathway. At this point, the HCC cell proliferation, invasion, and migration were attenuated, corresponding to facilitated cell apoptosis and repressed xenograft growth.
Materials and methods
Tissue specimens
A total of 24 cases of HCC tissues (lesion tissues pathologically identified as HCC) and matched adjacent normal tissues (> 2 cm from the lesion without any cancer cells) were obtained from patients who were diagnosed as HCC and received partial hepatectomy at the Xiangya Hospital from January 2018 to January 2019. The study was approved by the Institutional Review Board of Xiangya Hospital. Written informed consent was obtained from each participant. The fresh tissues were temporarily stored in liquid nitrogen and permanently preserved at −80ºC.
Short hairpin RNA targeting has_circ_0015756 or FAK
The sequence used to inhibit has_circ_0015756 or FAK expression was cloned into pENTR™/U6 (Invitrogen, USA). Sense oligos used were 5ʹ-CACCGCACCTCAATGCAAAGAAAAAACGAATTTTTTCTTTGCATTGAGGTG-3ʹ (has_circ_0015756) and 5ʹ- CACCGCAAGAACTTAGCAGGAAATGCGAACATTTCCTGCTAAGTTCTTGC-3ʹ (FAK). Antisense oligos used were 5ʹ-AAAACACCTCAATGCAAAGAAAAAATTCGTTTTTTCTTTGCATTGAGGTGC-3ʹ (has_circ_0015756) and 5ʹ-AAAAGCAAGAACTTAGCAGGAAATGTTCGCATTTCCTGCTAAGTTCTTGC-3ʹ (FAK). Sense oligos of scramble shRNA were 5ʹCACCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGACTTTTTTC-3ʹ and antisense oligos were 5ʹ-AAAAGAAAAAAGTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGACA-3ʹ.
Cell culture and cell treatment
A human fetal hepatic cell line (WRL-68 cells) and HCC cell lines (MHCC97H, Bel-7402, Huh-7 and Bel-100) (all purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China) were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 mg/ml streptomycin in a humidified atmosphere of 5% CO2. To construct HCC cells with circ_0015756 or FAK knockdown, cells were transfected with small hairpin RNA (Fenghuibio Inc., Changsha, Hunan) against circ_0015756 (sh-circ_0015756) and sh-FAK, respectively. To perturb miR-7 expression in HCC cells, cells were cultured with miR-7 mimic or miR-7 inhibitor (Shanghai GenePharma Co. Ltd., Shanghai, China). Cell treatment was performed using Lipofectamine 3000 as per the manufacturer’s specification (D0010, Beijing Solarbio Science & Technology Co., Ltd, China).
RNA isolation and quantification
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. For miRNA quantification, total RNA was reverse-transcribed by the TaqMan MicroRNA Reverse Transcription (RT) Kit (Applied Biosystems, Foster City, CA, USA) and then subjected to Taqman miRNA assay and TaqMan Universal PCR Master Mix (Applied Biosystems) according to the manufacturer’s protocol. The relative expression of miR-7 was normalized by the U6 transcript. For the quantification of circ_0015756, cDNA was synthesized by use of the EasyScript First-Strand cDNA Synthesis SuperMix kit (AE301-02, TransGen Biotech, Beijing, China) according to the manufacturer’s procedures, and quantitation was performed using SYBR®Premix Ex TaqTM II (RR820A, Takara Bio Inc., Dalian, China). A house-keeping gene, GAPDH, was used for normalization. The primer sequences are showed in . Quantitative polymerase chain reaction (qPCR) was run using the Applied Biosystems 7500 Real-Time PCR System with each reaction run in triplicate. Analysis and fold change were determined using the comparative threshold cycle (Ct) method [Citation22].
Table 1. The primer sequences for RT-qPCR
Protein extraction and western blot analysis
Total cell lysates were prepared using ice-cold radioimmunoprecipitation assay (RIPA) buffer (Sigma) containing phenylmethylsulfonyl fluoride (PMSF) (1 ml RIPA buffer was supplemented with 10 µl PMSF, both purchased from Beyotime Institute of Biotechnology, Shanghai, China). Western blotting analysis was performed and quantitated as described [Citation23]. Briefly, proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Beyotime Institute of Biotechnology). The membranes were incubated with anti-FAK antibody (1:1000, Cell Signaling Technology, Beverly, MA, USA), anti-Snail-1 antibody (1:1000, Cell Signaling Technology), anti-Slug antibody (1:800, Cell Signaling Technology), anti-MMP-9 antibody (1:1,000, ab73734, Abcam, Cambridge, MA, USA), anti-Akt antibody (1:500, ab8805), anti-p-Akt antibody (ab18206, 1μg/ml), and anti-GAPDH antibody (1:50, ab37168) overnight at 4℃. After the membranes were incubated with goat anti-rabbit immunoglobulin G (1:5,000, ab6721) at room temperature for 49 min, the bands were visualized and analyzed by the Image J software available from the NIH Web site. GAPDH was used for normalization. All antibodies were purchased from Abcam (Cambridge, MA, USA).
Cell viability assay
Cells were seeded into 96-well plates and then incubated with 20 μl MTT (5 mg/ml) at 37°C at the beginning, 24th, 48th and 72nd h, respectively. Four hours later, the media was aspirated and replaced by 150 μl Dimethyl Sulfoxide (DMSO, Amresco, Solon, Ohio, USA) in each well to dissolve the formazan crystals. Absorbance was read at 490 nm using a Microplate reader (DNM-9602G; Aolu Biotech, Shanhai, China).
Cell invasion assay
Cell invasion assays were carried out using a Matrigel-coated transwell invasion chamber (BD Biosciences, San Jose, CA, USA) following the manufacturer’s instruction. In brief, the cells were added to the upper chambers at a concentration of 2.5 × 104 cells per well. Dulbecco’s modified Eagle’s medium (DMEM) with 5% FBS used as a chemoattractant in the lower chambers. The cells that transferred to the bottom chamber were stained with 0.1% crystal violet at room temperature for 15 min and then counted in six fields per well using the Olympus inverted microscope (Olympus, Tokyo, Japan).
Cell migration assay
Cell migration was evaluated by using a scratch assay. In brief, cells were seeded in a 6-well plate (2 × 106 cells/well) and a pipette (1 ml) was used to create a straight scratch. After 24-hour incubation, the scratch healing was captured using the Olympus inverted microscope (Olympus, Tokyo, Japan) and the distance from one side of the scratch to the other was measured.
Cell apoptosis assay
An apoptosis assay was performed using an Annexin V-FITC/PI Cell Apoptosis kit (KeyGen, Nanjing, Jiangsu, China). Briefly, a suspension (100 µl) of 5 × 105 cells was incubated with 5 µl of Annexin V and 1 µl of propidium iodide (PI) at room temperature for 15 min. The apoptotic rate was determined by flow cytometry as per the manufacturer’s specification (BD Pharmingen, San Diego, CA, USA).
Dual-luciferase reporter assay
The pmirGLO-based luciferase reporter plasmids (Promega, Madisom, WI, USA) containing wild-type circ_0015756 (pmirGLO-circ_0015756-wt), circ_0015756 mutated at the putative miR-7 binding sites (pmirGLO-circ_0015756-mut), wild-type FAK (pmirGLO-FAK-wt) or FAK mutated at the putative miR-7 binding sites (pmirGLO-FAK-mut) were designed. HEK293T cells (Shanghai Beinuo Biotech Ltd., Shanghai, China) were seeded into 6-well plates in triplicate and co-transfected with well-designed pmirGLO-based reporter plasmids and either miR-7 mimic or inhibitor using the dual-luciferase reporter assay system according to the manufacturer’s instructions (D0010, Beijing Solarbio Science & Technology Co., Ltd, China). Relative Luciferase activity was normalized to that of Firefly Luciferase.
Animal studies
A total of 60 BALB/c nude mice (without limitation in gender, aged 4 weeks, weighing 18 to 25 g) were purchased from Guangdong Medical Laboratory Animal Center (Guangdong, China). The mice were fed in specific pathogen-free (SPF) environment and subcutaneously injected with MHCC-97H cells at 1 × 106 cells per mouse (n = 30). When the tumor size reached about 200–300 mm3, the mice were injected with either lentivirus packaging vectors pLenti7.3/V5-DEST (Invitrogen, USA) constructed with pENTR™/U6-sh-circ_0015756 or pENTR™/U6-scramble shRNA (a viral titre of 5 × 108 TU/ml and 300 μl for per mouse) by tail vein injection. Five days later, the mice were injected intraperitoneally with piperidazole at 60 mg/kg and then euthanized by carbon dioxide asphyxiation. For evaluation of intrahepatic metastasis, orthotopic implantation in 30 BALB/c nude mice was performed. The cell suspensions of MHCC-97H cells (200 ml/mouse) were implanted into the left lobe of mouse liver, and then the mice were also injected with either lentivirus packaging vectors pLenti7.3/V5-DEST (Invitrogen, USA) constructed with pENTR™/U6-sh-circ_0015756 or pENTR™/U6-scramble shRNA (a viral titre of 5 × 108 TU/ml and 300 ul for per mouse) by tail vein injection. These mice were euthanized on day 36 after tumor implantation and the liver lesion was subject to hematoxylin and eosin (HE) staining to observe the metastasis. All experimental procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and approved by our institutional ethical guidelines for animal experiments.
Statistical analysis
Statistical analyzes were conducted by SPSS 22.0 software (IBM Corp. Armonk, NY, USA). Measurement data were expressed by mean ± standard deviation (SD). First, normal distribution and homogeneity of variance are tested. If the data conformed to normal distribution and homogeneity of variance, data between two groups were compared by non-paired t test, comparisons among multiple groups were analyzed by one-way analysis of variance (ANOVA) or repeated measurement ANOVA. p < 0.05 indicated that the difference was statistically significant.
Results
Expression patterns of circ_0015756 and mir-7 in HCC cells
In order to determine the expression patterns of circ_0015756 and miR-7 in HCC, we quantified the expressions of circ_0015756 and miR-7 in HCC tissues and matched adjacent normal tissues. The results showed that the expression of circ_0015756 was up-regulated and the expression of miR-7 was down-regulated in HCC tissues in contrast to matched adjacent normal tissues (p < 0.05, ). In addition, we determined the expressions of circ_0015756 and miR-7 in WRL-68, MHCC97H, Bel-7402, Huh-7 and Bel-100 cells by RT-qPCR. The results showed that the expression of circ_0015756 was higher in all HCC cells than in WRL-68 cells, while the expression of miR-7 was lower in all HCC cells than in WRL-68 cells (p < 0.05, ). Huh7 and MHCC97H cells were selected for in vitro studies.
Figure 1. Expression patterns of circ_0015756 and miR-7 in HCC cells. The expressions of circ_0015756 (a) and miR-7 (b) in HCC tissues and matched adjacent normal tissues The expressions of circ_0015756 (c) and miR-7 (d) in WRL-68, Bel-100, Bel-7402, MHCC97H, and Huh-7 cells were quantified by RT-qPCR, normalized to GAPDH (for circ_0015756) and U6 (for miR-7). * p < 0.05, ** p < 0.01, *** p < 0.005 . The experiment was performed in triplicate and repeated three times. The data were expressed by mean ± SD and were analyzed by one-way ANOVA.
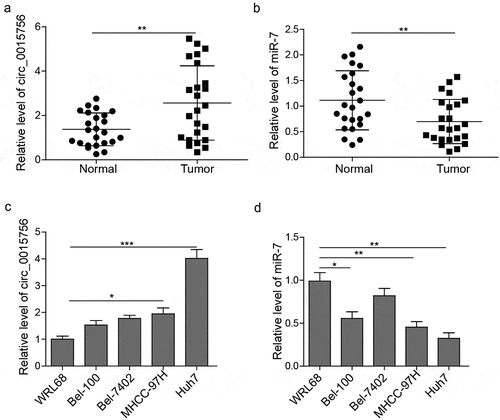
Circ_0015756 knockdown inhibited HCC cells in vitro
Following our speculation in regards to whether circ_0015756 had the potential to affect HCC progression, we prepared Huh7 and MHCC-97H cells transfected with either sh-circ_0015756 or scramble shRNA to examine the effects on HCC cells in vitro, focusing on cell viability, migration, invasion and apoptosis. To start, RT-qPCR was conducted to examine the expressions of circ_0015756 and miR-7 in Huh7 and MHCC-97H cells, and the results showed that disrupting circ_0015756 elevated the expression of miR-7 (p < 0.05, ). Next, the data obtained from the MTT assay, Annexin V-FITC/PI staining, scratch test and Transwell assay showed that cell viability, migration, invasion were reduced but the apoptosis was induced in Huh7 and MHCC-97H cells in the absence of circ_0015756 (p < 0.05, ). To further study the effect of circ_0015756 on FAK, Akt pathway, and HCC cell metastasis, we performed Western blot analysis to quantify the protein levels of FAK, p-Akt, Snail-1, Slug and MMP-9 once circ_0015756 was manipulated in Huh7 and MHCC-97H cells. Snail-1, Slug and MMP-9 genes are recognized to be involved in EMT and metastasis in cancers. As expected, the result showed the loss of circ_0015756 reduced expressions of FAK, Snail-1, Slug and MMP-9, and inhibited Akt pathway (p < 0.05, ). These findings suggested that circ_0015756 might be responsible for targeting miR-7 and inhibiting HCC cells.
Figure 2. Knockdown of circ_0015756 inhibits HCC cell viability, migration, and invasion but induces apoptosis in vitro. The levels of circ_0015756 (a) and miR-7 (b) in MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA circ_0015756 were determined by RT-qPCR, normalized to GAPDH (for circ_0015756) and U6 (for miR-7). (c) Cell viability of MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA circ_0015756 was evaluated by MTT assay. (d) Cell apoptosis of MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA circ_0015756 was performed by Annexin V-FITC/PI staining of flow cytometry. (e) Cell migration of MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA circ_0015756 was confirmed by scratch test. (f) Cell invasion of MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA circ_0015756 was performed by Transwell assay. (g)The protein levels of FAK, Akt, p-Akt, Snail-1, Slug and MMP-9 in MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA circ_0015756 were determined by Western blotting, normalized to GAPDH. * p < 0.05 and ** p < 0.01 vs. scramble shRNA (sh-NC). The experiments were performed in triplicate and repeated three times. The data were expressed by mean ± SD and were analyzed by one-way ANOVA in each panel, except panel C in which the data were analyzed by two-way ANOVA.
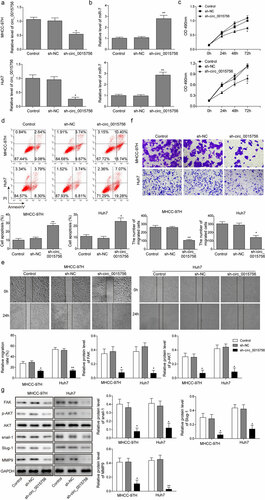
Mir-7 was found to inhibit HCC cells in vitro
With the results in the above section detailing circ_0015756 knockdown elevated the expression of miR-7, the focus of the experiment shifted to evaluate the effects of miR-7 on HCC cells in vitro. miR-7 mimic and inhibitor was introduced to establish Huh7 and MHCC-97H cells with or without miR-7. Firstly, we performed RT-qPCR and found miR-7 mimic or inhibitor could significantly increase or reduce miR-7 (p < 0.05, ). Subsequently, we conducted MTT assay, Annexin V-FITC/PI staining, scratch test and Transwell assay and observed that increased miR-7 decreased Huh7 and MHCC-97H cell viability, migration and invasion but induced apoptosis, while decreased miR-7 elicited opposite effects (p < 0.05, ). The next step in this process was to explore the mechanism by which miR-7 regulated HCC cells. We performed Western blot analysis and found the miR-7 overexpression resulted in reduced expressions of FAK, Snail-1, Slug and MMP-9, and inhibited Akt pathway, while miR-7 downexpression showed the opposite trend (p < 0.05, ). These findings suggested that miR-7 inhibited HCC cells in vitro, which triggered the hypothesis that FAK and Akt pathway functioned via miR-7.
Figure 3. miR-7 inhibits HCC cell viability, migration, and invasion but induces apoptosis in vitro. (a) The expression of miR-7 was determined by RT-qPCR, normalized to U6. (b) Cell viability of MHCC-97H and Huh-7 cells treated with miR-7 mimic or inhibitor was evidenced by MTT assay. (c) Cell apoptosis of MHCC-97H and Huh-7 cells treated with miR-7 mimic or inhibitor was performed by Annexin V-FITC/PI staining of flow cytometry. (d) Cell migration of MHCC-97H and Huh-7 cells treated with miR-7 mimic or inhibitor was determined by scratch test. (e) Cell invasion of MHCC-97H and Huh-7 cells treated with miR-7 mimic or inhibitor was performed by Transwell assay. (f) The expressions of FAK, Akt, p-Akt, Snail-1, Slug and MMP-9 in MHCC-97H and Huh-7 cells treated with miR-7 mimic or inhibitor were determined by Western blot, normalized to GAPDH. * p < 0.05, **p < 0.01. The experiments were performed in triplicate and repeated three times. The data were expressed by mean ± SD and were analyzed by one-way ANOVA in each panel, except panel B in which the data were analyzed by two-way ANOVA.
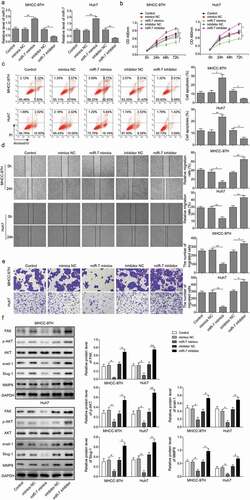
Knockdown of FAK inhibited HCC cells in vitro
To further confirm the mechanism by which miR-7 regulated HCC cells is dependent of FAK, Huh7 and MHCC-97H cells were treated with sh-FAK or scramble shRNA. By performing MTT assay, Annexin V-FITC/PI staining, scratch test and Transwell assay, we found that knockdown of FAK reduced Huh7 and MHCC-97H cell viability, migration and invasion but increased apoptosis (p < 0.05, ). The results of western blot analysis showed that knockdown of FAK reduced protein levels of Snail-1, Slug and MMP-9 and inhibited Akt pathway (p < 0.05, ). These findings suggested that FAK-mediated Akt-pathway was a potential anti-HCC mechanism.
Figure 4. Knockdown of FAK inhibits HCC cell viability, migration, and invasion but induces apoptosis in vitro. (a) Cell viability of MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA FAK was performed by MTT assay. (b) Cell apoptosis of MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA FAK was determined by Annexin V-FITC/PI staining of flow cytometry. (c) Cell migration of MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA FAK was evidenced by scratch test. (d) Cell invasion of MHCC-97H and Huh-7 cells manipulated by scramble shRNA and shRNA FAK was performed by Transwell assay. (e) The expressions of FAK, Akt, p-Akt, Snail-1, Slug and MMP-9 were determined by Western blotting, normalized to GAPDH. * p < 0.05 and ** p < 0.05 vs. scramble shRNA (sh-NC). The experiment was performed in triplicate and repeated three times. The data were expressed by mean ± SD and were analyzed by one-way ANOVA in each panel, except panel A in which the data were analyzed by two-way ANOVA.
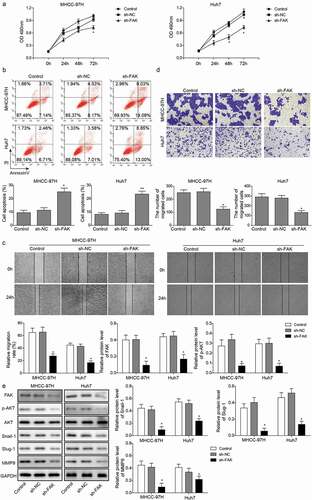
The interference of mir-7 restored HCC proliferation in the absence of circ_0015756
We used a computer-based miR target detection programs to predict the binding sites of circ_0015756 and miR-7 (http://starbase.sysu.edu.cn/), and miR-7 and FAK (http://www.targetscan.org/vert_72/), and the results were positive (). To determine whether circ_0015756 could bind with miR-7, the dual-luciferase reporter assay using vectors constructed with circ_0015756 sequences with recognizing sites or recognizing sites mutated in the presence of miR-7 mimic or inhibitor was performed. Decreases in Renilla luciferase were measured upon miR-7 mimic and pmirGLO-circ_0015756-wt treated and normalized to Firefly luciferase (p < 0.05, ). To determine whether the FAK gene is indeed the target of miR-7, dual-luciferase reporter vectors were constructed containing the predicted seed sequence in the 3ʹ-untranslated regions (3ʹUTR) of FAK, as well as the accordingly mutant vectors in which random nucleotide mutants were introduced into the seed sequences. Following the treatment with miR-7 mimic and vectors containing the FAK 3ʹ-UTR with recognizing sites, the Renilla luciferase was reduced and normalized to Firefly luciferase (p < 0.05, ). These findings suggested that circ_0015756 might be responsible for the dysregulation of miR-7 that negatively regulated FAK. Next, we performed recovery experiments to confirm that miR-7 inhibitor could reverse the effects of shcirc_0015756 on Huh7 and MHCC-97H cells, with sh-NC + NC inhibitor, shcirc_0015756 + NC inhibitor, sh-NC + miR-7 inhibitor and shcirc_0015756 + miR-7 inhibitor grouped. The results of RT-qPCR showed that the interference of miR-7 by its specific miRNA inhibitor was ablated by shcirc_0015756 (p < 0.05, ). The MTT assay (), Annexin V-FITC/PI staining (), scratch test () and Transwell assay () demonstrated that the interference of miR-7 restored the viability, migration and invasion and reduced the apoptosis of circ_0015756-depleted Huh7 and MHCC-97H cells. The results of Western blot analysis () showed that compared with the sh-NC + miR-7 inhibitor group, the shcirc_0015756 + miR-7 inhibitor exhibited reduced expressions of FAK, Snail-1, Slug and MMP-9, and inhibited Akt pathway, suggesting the interference of miR-7 removed the inhibitory effect of FAK, Snail-1, Slug and MMP-9, and Akt pathway.Circ_0015756 knockdown suppressed tumor growth and intrahepatic metastais of HCC
Figure 5. Interactions between circ_0015756 and miR-7, miR-7 and FAK. (a) The binding sites of circ_0015756 and miR-7. (b) The binding sites of miR-7 and FAK. (c) Luciferase activity of luciferase reporter vectors constructed with circ_0015756 sequences with recognizing sites or recognizing sites mutated in the presence of miR-7 mimic or inhibitor. (d) Luciferase activity of luciferase reporter vectors containing the FAK 3ʹ-UTR with recognizing sites or recognizing sites mutated. (e) The expression level of miR-7 in MHCC-97H and Huh-7 cells was determined by RT-qPCR, normalized to GAPDH. (f) Cell viability of MHCC-97H and Huh-7 cells was examined by MTT assay. (g) Cell apoptosis of MHCC-97H and Huh-7 cells was examined by Annexin V-FITC/PI-labeled flow cytometry. * p < 0.05, **p < 0.01. The experiments were performed in triplicate and repeated three times. The data were expressed by mean ± SD and were analyzed by one-way or two-way ANOVA.
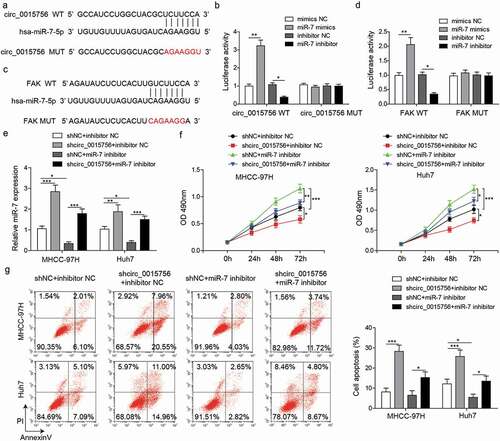
Figure 6. Knockdown of circ_0015756 inhibits HCC cell migration and invasion, FAK expressiom and Akt pathway by binding to miR-7. (a) Cell migration of MHCC-97H and Huh-7 cells was evaluated by scratch test. (b) Cell invasion of MHCC-97H and Huh-7 cells was evaluated by Transwell assay. (c) The expressions of FAK, Akt, p-Akt, Snail-1, Slug and MMP-9 were determined by Western blotting, normalized to GAPDH. * p < 0.05, **p < 0.01. The experiments were performed in triplicate and repeated three times. The data were expressed by mean ± SD and were analyzed by one-way ANOVA.
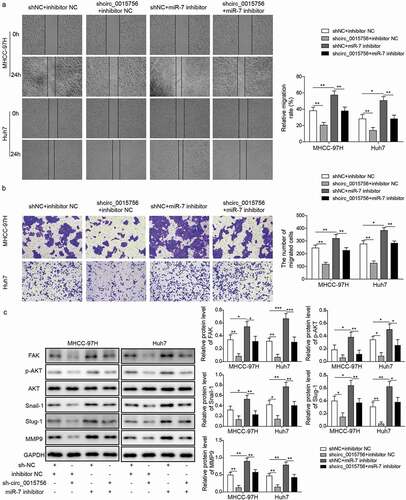
Figure 7. Knockdown of circ_0015756 suppresses tumor growth and intrahepatic metastasis of HCC in vivo. a-c, Thirty BALB/c nude mice were subcutaneously injected with MHCC-97H cells at 1 × 106 cells per mouse. When the tumor size reached about 200–300 mm3, the mice were injected with either lentivirus particles containing sh-circ_0015756 (sh-circ_0015756 group, n = 10) or scramble shRNA (sh-NC group, n = 10). The control group consisted of 10 mice bearing tumor xenografts of MHCC-97H cells without treatment of lentivirus particles. (a) Representative subcutaneous xenotransplanted tumors. (b) Tumor volume. (c) Tumor weight. D-F, The cell suspensions of MHCC-97H cells were implanted into the left lobe of mouse liver for orthotopic implantation. (d) Representative liver lesions in mice after orthotopic implantation (n = 10 for each group). (e) Representative views of liver lesions by HE staining (n = 10 for each group). (f) Number of metastasis nodules (n = 10 for each group). * p < 0.05 and *** p < 0.05 vs. scramble shRNA (sh-NC). The experiments were performed in triplicate and repeated three times. The data were expressed by mean ± SD and were analyzed by one-way or two-way ANOVA.
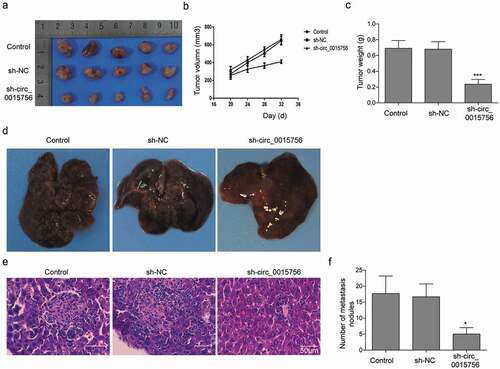
Finally, the finding that circ_0015756 knockdown inhibited HCC cells in vitro was reproduced in tumor xenografts of HCC cells. Thirty BALB/c nude mice were subcutaneously injected with SMCC7721 cells at 1 × 106 cells per mouse. When the tumor size reached about 200–300 mm3, the mice were injected with either lentivirus particles containing sh-circ_0015756. The results showed that the mice bearing tumor xenografts of SMCC7721 cells showed reduced tumor volume () and tumor weight () after injection with lentivirus particles containing sh-circ_0015756 relative to injection with lentivirus containing scramble shRNA or without injection of lentivirus particles (p < 0.05). Next, to study the effect of circ_0015756 on intrahepatic metastasis of HCC, orthotopic implantation was performed in 30 BALB/c nude mice. As depicted in , circ_0015756 knockdown suppressed intrahepatic metastasis of HCC.
Discussion
CircRNAs represent a large class of endogenous, non-coding RNAs, which function in cellular physiology with such distinct properties as high stability, and availability in body fluids, holding promises as molecular diagnostic biomarkers, non-invasive monitors, prognostic factors, and therapeutic targets [Citation24]. The growing studies have expanded the number of circRNAs with aberrant expression in HCC [Citation25–Citation27]. For example, the highly expressed circRNA_100338 in HCC has been validated to predict an unfavorable survival rate and facilitated metastatic potential [Citation28]. Importantly, the obtained results of this current study suggested that the expression of circ_0015756 was significantly higher in HCC cells than in normal WRL-68 cells. It is important to note that a previous expression profiling study identified that circ_0015756, upregulated in HCC cells, was proposed as a miR-1250-3p sponge to mediate hepatoblastoma progression [Citation9]. In this study, we further investigated the involvement of circ_0015756 in HCC progression, and delineated that knockdown of circ_0015756 inhibited HCC cell viability, migration, and invasion but induced apoptosis in vitro. Additionally, circ_0015756 knockdown suppressed the tumor formation abilities of HCC cells in vivo.
It has been established that circRNAs are involved in the modulation of liver homeostasis and disease, due to their roles in controlling the growth of tissues, inflammatory responses and cell cycle and apoptosis [Citation29]. Several circRNAs act as miRNA sponges to regulate the expression of target genes, thus playing a key role in the HCC progression [Citation30]. Besides, miRNAs functions in gene-regulation events via pairing to the mRNAs of protein-coding genes to repress their expression [Citation31]. In this study, up-regulation of circ_0015756 expression might be responsible for the dysregulation of miR-7 that negatively regulated FAK. Dual-luciferase reporter assay confirmed that circ_0015756 could bind to miR-7, and FAK gene was indeed a target gene of miR-7. A recent study also reported a circRNA-miRNA-gene axis that hsa_circ_0005075 mediated miRNAs including miR-23b-5p, miR-93-3p, miR-581, miR-23a-5p and their targeted mRNAs, involving in cell adhesion during HCC development [Citation32]. In addition, a network of circRNA-miRNA-mRNA has been explored in HCC, the findings of which indicated that circRNA-101,368 directly bound to miR-200a to attenuate the migration of HCC cells by regulating high mobility group box 1 (HMGB1) [Citation33]. Moreover, another upregulated circRNA in HCC tissues, ciRS-7, was found to sponge miR-7, thus augmenting the HCC development [Citation16]. Based on our findings and those studies, we could assume a circ_0015756/miR-7/FAK axis may participate in the regulation of HCC progression.
This study revealed that the expression of miR-7 was significantly downregulated in HCC cells. Consistently, a previously conducted study also provided evidence suggesting that miR-7 was downregulated in HCC tissues versus the adjacent normal tissues [Citation34]. The gain-of-function experiments in this study subsequently showed that the upregulation of miR-7 inhibited HCC cell viability, migration, and invasion but induced apoptosis in vitro. Mounting evidence verified miR-7 as an anti-tumor miRNA in human HCC through targeting and inhibiting the expression of oncogenes [Citation13,Citation35]. As a new functional target of miR-7, FAK was found to be an oncogene in HCC. Our study proved that knockdown of FAK repressed HCC cell viability, migration, and invasion but induced apoptosis in vitro. A study has suggested that FAK is a target gene of miR-7, and importantly, rescue of FAK expression could reverse the inhibitory role of miR-7 in cell migration and invasion of breast cancer [Citation18]. On the basis of the study of Luedde T, FAK could enhance HCC cell motility and spreading by interacting synergistically with miR-151 [Citation36]. Additionally, a newly identified cirRNA circFNDC3B, which is related to invasion, achieved the tumor suppressive role through inactivating the SRC/FAK signaling pathway by sponging miR-1178-3p in bladder cancer [Citation37]. Furthermore, the present study demonstrated that FAK-mediated Akt-pathway was a potential mechanism of miR-7 inhibiting the progression of HCC. In a study conducted by Fang et al., miR-7 efficiently blocked the PI3K/Akt pathway, thus suppressing the tumorigenesis [Citation38]. FAK was revealed to activate the Akt signaling pathway during the lung metastases in hepatocellular carcinoma [Citation39]. Besides, in response to the activation of the FAK/Akt pathway stimulated by miR-6875-3p, the tumorigenesis of HCC in vitro and in vivo was robustly enhanced [Citation40]. It has been well recognized that Snail, Slug and MMPs have significant roles to play in the cancer progression by facilitating EMT and invasion, including HCC [Citation41,Citation42]. The activation of the PI3K/Akt/Snail1/E-cadherin axis was shown to cause significant increase in breast cancer cell viability and migration [Citation43]. The anti-cancer effects could be realized by inhibiting the PI3K/Akt pathway in HCC, as reflected by reduced MMP-9 expression [Citation44]. Consistently, promoted expression of Slug through the Akt pathway was shown to accelerate the EMT of hepatoma cells and ultimately the malignancy of HCC [Citation45]. As a result, a network of miR-7/FAK/Akt may underlie the regulatory role of circ_0015756 in HCC.
Taken together, the study demonstrates that the konckdown of circ_0015756 leads to impeding the HCC development both in vitro and in vivo. Our study presents a comprehensive understanding of molecular mechanism in which circ_0015756 sponge miR-7 to upregulate the expression of FAK, promoting the Akt pathway. This finding reveals novel insights into mechanisms of circ_0015756-induced carcinogenicity in HCC, and mRNA, miRNA, circRNA and pathway are studied. However, due to the effectiveness of multiple forms of intervention has not been clearly determined, in this study, only shRNAs, mimics or inhibitors were applied to perturb the expression of involved mRNA, miRNA and circRNA. Therefore, more effective and specific methods perturbing those targets warrant to be exploited.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Sergio A, Cristofori C, Cardin R, et al. Transcatheter arterial chemoembolization (TACE) in hepatocellular carcinoma (HCC): the role of angiogenesis and invasiveness. Am J Gastroenterol. 2008;103(4):914–921.
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255.
- Andrisani OM, Studach L, Merle P. Gene signatures in hepatocellular carcinoma (HCC). Semin Cancer Biol. 2011;21(1):4–9.
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314.
- He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144.
- Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565.
- Du WW, Zhang C, Yang W, et al. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7(17):4183–4191.
- Liu BH, Zhang BB, Liu XQ, et al. Expression profiling identifies circular RNA signature in hepatoblastoma. Cell Physiol Biochem. 2018;45(2):706–719.
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222.
- Li J, Qiu M, An Y, et al. miR-7-5p acts as a tumor suppressor in bladder cancer by regulating the hedgehog pathway factor Gli3. Biochem Biophys Res Commun. 2018;503(3):2101–2107.
- Luo J, Li H, Zhang C. MicroRNA-7 inhibits the malignant phenotypes of nonsmall cell lung cancer in vitro by targeting Pax6. Mol Med Rep. 2015;12(4):5443–5448.
- Kabir TD, Ganda C, Brown RM, et al. A microRNA-7/growth arrest specific 6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma. Hepatology. 2018;67(1):216–231.
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612.
- Yu L, Gong X, Sun L, et al. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7):e0158347.
- Xu L, Zhang M, Zheng X, et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17–27.
- Wu W, Liu S, Liang Y, et al. MiR-7 inhibits progression of hepatocarcinoma by targeting KLF-4 and promises a novel diagnostic biomarker. Cancer Cell Int. 2017;17:31.
- Kong X, Li G, Yuan Y, et al. MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS One. 2012;7(8):e41523.
- Ko BS, Jan YJ, Chang TC, et al. Upregulation of focal adhesion kinase by 14-3-3epsilon via NFkappaB activation in hepatocellular carcinoma. Anticancer Agents Med Chem. 2013;13(4):555–562.
- Chen JS, Huang XH, Wang Q, et al. Sonic hedgehog signaling pathway induces cell migration and invasion through focal adhesion kinase/AKT signaling-mediated activation of matrix metalloproteinase (MMP)-2 and MMP-9 in liver cancer. Carcinogenesis. 2013;34(1):10–19.
- Spangle JM, Roberts TM, Zhao JJ. The emerging role of PI3K/AKT-mediated epigenetic regulation in cancer. Biochim Biophys Acta Rev Cancer. 2017;1868(1):123–131.
- Chabot S, Orio J, Castanier R, et al. LNA-based oligonucleotide electrotransfer for miRNA inhibition. Mol Ther. 2012;20(8):1590–1598.
- Rojo AI, Medina-Campos ON, Rada P, et al. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: role of glycogen synthase kinase-3. Free Radic Biol Med. 2012;52(2):473–487.
- Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005.
- Qiu L, Huang Y, Li Z, et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13(2):441–455.
- Zhang X, Xu Y, Qian Z, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9(11):1091.
- Weng Q, Chen M, Li M, et al. Global microarray profiling identified hsa_circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet. 2019;56(1):32–38.
- Huang XY, Huang ZL, Xu YH, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7(1):5428.
- Song M, Xia L, Sun M, et al. Circular RNA in Liver: health and Diseases. Adv Exp Med Biol. 2018;1087:245–257.
- Zhou H, Xu Q, Ni C, et al. Prospects of Noncoding RNAs in Hepatocellular Carcinoma. Biomed Res Int. 2018;2018:6579436.
- Guo H, Ingolia NT, Weissman JS, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840.
- Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore). 2016;95(22):e3811.
- Li S, Gu H, Huang Y, et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle. 2018;17(19–20):2349–2359.
- Ma C, Qi Y, Shao L, et al. Downregulation of miR-7 upregulates Cullin 5 (CUL5) to facilitate G1/S transition in human hepatocellular carcinoma cells. IUBMB Life. 2013;65(12):1026–1034.
- Zhang X, Hu S, Zhang X, et al. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2014;443(3):1078–1084.
- Luedde T. MicroRNA-151 and its hosting gene FAK (focal adhesion kinase) regulate tumor cell migration and spreading of hepatocellular carcinoma. Hepatology. 2010;52(3):1164–1166.
- Liu H, Bi J, Dong W, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17(1):161.
- Fang Y, Xue JL, Shen Q, et al. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55(6):1852–1862.
- Leng C, Zhang ZG, Chen WX, et al. An integrin beta4-EGFR unit promotes hepatocellular carcinoma lung metastases by enhancing anchorage independence through activation of FAK-AKT pathway. Cancer Lett. 2016;376(1):188–196.
- Xie Y, Du J, Liu Z, et al. MiR-6875-3p promotes the proliferation, invasion and metastasis of hepatocellular carcinoma via BTG2/FAK/Akt pathway. J Exp Clin Cancer Res. 2019;38(1):7.
- Qiao B, Johnson NW, Gao J. Epithelial-mesenchymal transition in oral squamous cell carcinoma triggered by transforming growth factor-beta1 is Snail family-dependent and correlates with matrix metalloproteinase-2 and −9 expressions. Int J Oncol. 2010;37(3):663–668.
- Song FN, Duan M, Liu LZ, et al. RANKL promotes migration and invasion of hepatocellular carcinoma cells via NF-kappaB-mediated epithelial-mesenchymal transition. PLoS One. 2014;9(9):e108507.
- Jin X, Zhu L, Cui Z, et al. Elevated expression of GNAS promotes breast cancer cell proliferation and migration via the PI3K/AKT/Snail1/E-cadherin axis. Clin Transl Oncol. 2019.
- Zhu P, Liu Z, Zhou J, et al. Tanshinol inhibits the growth, migration and invasion of hepatocellular carcinoma cells via regulating the PI3K-AKT signaling pathway. Onco Targets Ther. 2019;12:87–99.
- Huan HB, Yang DP, Wen XD, et al. HOXB7 accelerates the malignant progression of hepatocellular carcinoma by promoting stemness and epithelial-mesenchymal transition. J Exp Clin Cancer Res. 2017;36(1):86.
