ABSTRACT
Lung cancer (LC) is one of the malignant tumors with growing morbidity and mortality. The involvement of runt-related transcription factor 1 (RUNX1) in LC patients has been elucidated. We intended to research mechanisms of RUNX1 and tartrate-resistant acid phosphatase 5 (ACP5) in LC. Firstly, ACP5 levels in LC tissues, paracancerous tissues, LC cells and tracheal epithelial cells were detected. RUNX1 overexpression plasmid and interference plasmid were constructed and transfected into 95C cells and A549 cells, respectively. The binding of RUNX1 to ACP5 promoter was tested. Additionally, the gain- and loss-of-function were performed to explore the effects of ACP5 and RUNX1 on LC biological process. The xenograft tumor in nude mice was constructed in vivo to verify in vitro results. Functional rescue experiment was performed by adding MAPK-specific activator P79350 to A549 cells with si-ACP5 to measure the effects of ERK/MAPK axis on LC progression. Consequently, we found ACP5 expression was higher in LC tissues and cells, and ACP5 silencing suppressed LC cell growth. Overexpression of ACP5 promoted malignant biological behavior of LC cells. RUNX1 could bind to ACP5 promoter, and overexpressed RUNX1 promoted ACP5 expression and LC cell growth. Moreover, ACP5 upregulated the ERK/MAPK axis and thus promoted LC progression. The results of xenograft tumor in nude mice showed that silencing ACP5 could inhibit the growth of LC cells in vivo. To conclude, silenced RUNX1 inhibits LC progression through the ERK/MAPK axis by binding to ACP5. This study may provide new approaches for LC treatment.
1. Introduction
Lung cancer (LC) has long been regarded as the most concerned malignancy in worldwide, contributing to about 1.8 million newly diagnosed patients, and 1.6 million deaths annually, with a 5-year survival rate of 4 ~ 17% [Citation1]. Majority of new cases are non-small cell lung cancer (NSCLC), of which over half are regarded as locally advanced [Citation2] and unfortunately, the five-year survival rate is only 16% [Citation3]. Smoking is identified to be the major cause of LC, which is responsible for over 80% cases [Citation4]. Additionally, air pollution, radon and radiation exposure, hereditary susceptibility, unbalanced diet are increasingly recognized as the risk factors for LC [Citation5]. At present, treatment options for LC are mainly surgical resection, radiotherapy, chemotherapy and targeted therapies, alone or in combination, varying from tumor location and stages [Citation6]. However, most newly diagnosed LC patients are unresectable and resistant to chemotherapy and radiotherapy [Citation7]. Therefore, there is an urgent need to find valuable biomarkers in the early diagnosis for prevention and effective treatments of LC patients.
Runt-related transcription factor family (RUNX), including RUNX1, RUNX2 and RUNX3, could exert functions in embryological development, nervous system, angiogenesis, hematopoiesis and immune responses [Citation8]. It is well recognized that RUNX1 mutation is associated with poor clinical outcomes in patients with acute myeloid leukemia [Citation9]. Importantly, RUNX1 is highly expressed in mesenchymal and epithelial cells of the lung during development and postnatal period [Citation10]. However, there are few researches about the correlation between RUNX1 and LC. In this study, we found RUNX1 could bind to tartrate-resistant acid phosphatase 5 (ACP5) and promote LC cell growth. ACP5 is derived from osteoclasts, osteoblasts and osteocytes [Citation11]. Interestingly, high ACP5 expression is associated with tumor development and serves as an underlying prognostic marker for lung adenocarcinoma [Citation12]. Besides, the extracellular signal regulated kinase (ERK)/mitogen activated protein kinase (MAPK) pathway could monitor cellular biological processes in tumor initiation and progression by affecting the activation of various downstream molecules, varying from morphogenesis, differentiation, cell cycle distribution, apoptosis, metabolism to angiogenesis [Citation13]. Activated ERK/MAPK pathway in lung mesenchymal layers efficiently integrates epithelial–mesenchymal interactions to trigger lung development [Citation14]. Surprisingly, the activity of RUNX1 could be regulated by ERK1/2 after translation [Citation15]. From all the above, we hypothesized that RUNX1 and ACP5 might co-work in LC progression via the ERK/MAPK pathway. Thus, we conducted a study to investigate how RUNX1 co-works with ACP5 in LC by regulating the ERK/MAPK pathway to find a new clinical reference for LC patients.
2. Materials and methods
2.1. Ethics statement
This study was approved and supervised by the ethics committee of the Cancer Hospital Affiliated to Harbin Medical University. All the subjects signed the informed consent. The protocol was also approved by the Institutional Animal Care and Use Committee of the Cancer Hospital Affiliated to Harbin Medical University. Significant efforts were made to minimize the number of animals and their respective suffering.
2.2. Sample collection
From September 2012 to June 2013, 80 resected LC tissues and paracancerous tissues (over 5 cm away from cancer tissues) from LC patients (57 males and 23 females, median age: 49 ~ 79 years, average age: 64.9 ± 7.3 years) who were diagnosed and treated in the Cancer Hospital Affiliated to Harbin Medical University were enrolled in this experiment. All patients were followed up every 3 months for 5 years. Patients were enrolled into this study if they met the following criteria: 1) patients were diagnosed as LC by the detection of various indicators; 2) patients did not receive radiotherapy or chemotherapy; 3) patients had complete clinical data. Patients complicated with chronic system diseases or other malignant tumors would be excluded.
2.3. Cell culture
Human LC cell lines H1299, 95C, A549, SPC-A-1 and L9981, and human tracheal epithelial cells 16HBE and MRC-5 (Cancer Cell Bank of Chinese Academy of Medical Sciences, Beijing, China) were cultured at 1 × 105 cells/cm2 respectively in Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% fetal bovine serum (FBS) (Gibco Company, Grand Island, NY, USA) for 48 h (37°C, 5% CO2). When cell confluence got to 80% ~ 90%, cells were detached with 0.025% trypsin (Gibco Company, Grand Island, NY, USA) and passaged.
2.4. Cell transfection
The well-grown 95C cells were transfected with phosphate buffer saline (PBS), negative control (NC), or ACP5 overexpression (OE) plasmids, and, respectively, named as blank group, NC group and OE-ACP5 group. The well-grown A549 cells were transfected with PBS, NC, or ACP5 small interfering RNA (si-RNA), and, respectively, named as blank group, NC group and si-ACP5 group. In order to further verify the effects of RUNX1 on LC cell growth, the well-grown 95C cells were transfected with PBS, NC and OE-RUNX1 plasmids, and recorded as blank group, NC group and OE-RUNX1 group, respectively. The well-grown A549 cells were transfected with PBS, NC, or si-RUNX1, and, respectively, named as blank group, NC group and si-RUNX1 group. Then, ACP5 overexpression plasmid was constructed and transfected into A549 cells in the si-RUNX1 group, which was recorded as the RUNX1-ACP5 group. Overexpression plasmids and interfering plasmids used in the experiment were synthesized by and purchased from GenePharma (Shanghai, China). All operations were carried out as per the instructions of LipofectamineTM 3000 kit (Invitrogen, Carlsbad, CA, USA).
2.5. Reverse transcription quantitative polymerase chain reaction (rt-qpcr)
Total RNA from LC tissues and cells were obtained using the RNAiso Plus (Takara, Otsu, Shiga, Japan) and Trizol LS Reagent (Takara, Otsu, Shiga, Japan) separately. Then, formaldehyde denaturation electrophoresis was used to verify the reliability of obtained RNA, and subsequent experiments were continued. RT-PCR was conducted as per the manufacturer’s protocol using PrimeScript™ RT reagent kit (Takara, Otsu, Shiga, Japan). The mRNA expression was quantified by standard real-time qPCR protocol with SYBR Premix Ex Taq (Takara, Otsu, Shiga, Japan) with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a reference. The primers are shown in .
Table 1. Primer sequences of RT-qPCR
2.6. Western blot analysis
The total proteins were extracted by radio-immunoprecipitation assay2 lysis buffer containing phenylmethylsulfonyl fluoride (Beyotime Biotechnology Co., Ltd, Shanghai, China) and protein level in the supernatant was determined by bicinchoninic acid method. An equal volume (50 mg) of protein was loaded into 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and then transferred into the polyvinylidene fluoride membranes (Millipore Corp, Billerica, MA, USA). PVDF membranes were then incubated with tris-buffered saline tween (TBST) (Boster Biological Technology Co., Ltd, Wuhan, Hubei, China) containing 5% skimmed milk at room temperature to block nonspecific binding. Subsequently, the membranes were cultured with primary antibodies () at 4°C overnight, and then with rabbit anti-rat secondary antibodies at room temperature for 1 h. The proteins were colored in enhanced chemiluminescence reagent, and visualized using BioSpectrum gel imaging system (Bio-Rad, Hercules, CA, USA).
Table 2. Antibodies used for western blot analysis
2.7. Chromatin immunoprecipitation (chIP) assay
ChIP kit (ab500, Abcam, Cambridge, MA, USA) [Citation16] was used to detect the binding of RUNX1 protein to ACP5 promoter. All operations were carried out strictly as per the instructions.
2.8. Dual luciferase reporter gene assay
The binding relationship between RUNX1 and ACP5 promoter was validated by Dual-Glo® Double Luciferase Reporter Gene Detection System, E2920, Promega Corporation, Madison, Wisconsin, USA [Citation17]. All experimental operations were carried out strictly according to the instructions.
2.9. 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay
MTT cell proliferation and cytotoxicity assay kit (C0009, Beyotime Biotechnology Co., Ltd, Shanghai, China) was applied to detect cell viability. All operations were carried out strictly in accordance with the instructions.
2.10. Flow cytometry
The well-grown cells were washed with PBS, detached with 0.025% trypsin, and then incubated in Dulbecco’s Modified Eagle’s Medium (DMEM) with 2 mmol carboxyfluorescein diacetate succinimidyl ester for 15 ~ 30 min. After 3 PBS washes and centrifugation, the supernatant was discarded and cells were resuspended in DMEM. Finally, cell proliferation was measured by using a flow cytometer.
The cells were treated with 70% formaldehyde (v/v) at −20°C for 24 h. Next, cells were stained with 50 μg/mL propidium iodide (PI) containing 10 μg/mL RNase for 20 min. Finally, cell cycle distribution was measured using a flow cytometer.
Cell suspension was added with 5 μL Annexin V-fluorescein isothiocyanate (FITC) and 5 μL PI in the dark for 10 min. After staining, cell apoptosis was measured by a flow cytometer.
2.11. Immunofluorescence assay
Cells in the cover glass were washed in PBS 3 times and fixed in 4% polyformaldehyde for 15 min at 4°C. After treatment with 0.5% Triton-100 X for 20 min, the primary antibodies proliferating cell nuclear antigen (PCNA, 1: 200, ab92552) and Cytochrome C (1: 100, ab133504) were added to incubate cells overnight at 4°C. Afterward, cell slides were washed in PBS and incubated with Alexa Fluora 594 labeled secondary antibody fluorescent goat anti-rabbit antibody (1: 5000, ab150088, all from Abcam Inc., Cambridge, MA, USA) at 37°C for 1 h. Then, the nuclei were counterstained with 4ʹ, 6-diamidino-2-phenylindole (DAPI), and cells were observed under the fluorescence microscope (DM3000, Leica, Solms, Germany). MitotrackerTM Red CMXRos-Special Packaging (M7512, Thermo Fisher, Carlsbad, California, USA) was used for mitochondrial staining. All operations were performed strictly according to the instructions.
2.12. PI staining
Cells in each group were taken and fixed with 4% paraformaldehyde for 20 min, then stained with PI (3 μM) in the dark for 5 min. Finally, cells were analyzed using a fluorescence microscope. Five regions were randomly selected in each group for analysis. The apoptotic nuclei were stained red.
2.13. Xenograft tumors in nude mice
Thirty specific pathogen-free BALB/c nude mice (4 ~ 6 week-old, 20 ± 2 g) (Beijing Vital River Laboratory Animal Technology Co., Ltd, Beijing, China, SCXK (Beijing) 2015–0001), were numbered with body weight as a parameter and randomly assigned into six groups, with 5 mice in each group. Tumor volume was measured every 7 days after cell injection, mice were euthanized 28 days later, and tumors were taken out and weighed for immunohistochemistry. The tumor volume = M12 × M2 × 0.5236 [Citation18], in which M1 represented the shortest axis and M2 represented the longest axis. The tumor volume on day 0 was recorded as 0 mm3. After 28 days of cell injection, mice were euthanized and tumors were removed for weighing and histological experiments. The xenograft tumor tissues of mice in each group were extracted, and sliced into sections. Five sections were selected randomly and equidistantly for immunohistochemistry.
2.14. Immunohistochemistry
After routine treatment [Citation19], the sections were incubated with Ki67 antibody (1: 500, ab15580) and Caspase-3 (1: 1000, ab13847) (Abcam Inc., Cambridge, MA, USA) for 30 min. After 3 PBS washes, sections were dripped with 40 μL horseradish peroxidase-labeled streptavidin-working solution and incubated at 37°C for 15 min. Then, sections were washed with PBS 3 times again, visualized with 2, 4-diaminobutyric acid, counterstained with hematoxylin for 30 s after washing with distilled water, and sealed with neutral gum after dehydration. Five non-overlapping visual fields were selected in each section for observation under the microscope. The cells with brown-yellow or brown granules in the nucleus were Ki67 or Caspase-3 positive cells. Five regions were selected at random in each section to sum up the ratio of positive cells.
2.15. Statistical analysis
SPSS 21.0 (IBM Corp., Armonk, NY, USA) was applied for data analysis. Kolmogorov–Smirnov test showed whether the data were in normal distribution. The results were exhibited in mean ± standard deviation. The t test was employed for analysis of comparisons between two groups, one-way analysis of variance (ANOVA) for comparisons among multi-groups, and Tukey’s post hoc test for pairwise comparisons after ANOVA. Survival curve was drawn using Kaplan-Meier survival analysis, and the log rank test was used for post-analysis. p value was obtained by two-tailed test and p < 0.05 indicated significant difference.
3. Results
3.1. ACP5 is highly expressed in LC tissues and cells
According to the analysis of online database (https://www.proteinatlas.org/), ACP5 was markedly overexpressed in lung tissues ()). A previous study demonstrated that ACP5 was notably overexpressed in patients with hepatocellular carcinoma and could promote the growth of hepatocellular carcinoma cells [Citation20]. ACP5 mRNA expression in LC tissues of 80 LC patients was significantly higher than that in paracancerous tissues ()). The relationship between ACP expression and clinical characteristics of LC patients was analyzed. Patients whose relative ACP5 expression was lower than 5.53 (median value) were treated as low expression group, otherwise the high expression group. In LC tissues, ACP5 expression was related to clinical stage, lymph node metastasis, and tumor differentiation. ACP5 expression increased with the increase of clinical stage of LC patients. ACP5 expression in cancer tissues of LC patients at stage III + IV, patients with low differentiation, or with lymph node metastasis was notably higher than those at stage I + II, with high differentiation, or without lymph node metastasis (all p < 0.05). ACP5 expression was not statistically correlated with age, gender and tumor size (all p > 0.05) (). Subsequently, Kaplan-Meier survival analysis based on follow-up records revealed that LC patients with high ACP5 expression had worse prognosis, with a 5-year survival rate of 15.0% and an average survival time of 27.1 months after diagnosis, while patients with low ACP5 expression had a relatively better prognosis, with a 5-year survival rate of 30.0% and an average survival time of 40.4 months after diagnosis (all p < 0.05, )).
Table 3. Association between ACP5 expression with clinical characteristics of LC patients
Figure 1. ACP5 is highly expressed in LC tissues and cells. a. Relative ACP5 expression in different cancer tissues; b. Relative ACP5 expression in LC tissues and paracancerous tissues, *** p < 0.001, compared with paracancerous tissues; c. Kaplan-Meier survival analysis of LC patients with high or low ACP5 expression; d. Relative ACP5 expression in LC cells and tracheal epithelial cells, * p < 0.05, ** p < 0.01, compared with 16HBE cells; e. Relative ACP5 mRNA expression in 95C and A549 cells after different transfection, * p < 0.05, ** p < 0.01, compared with the NC group; f. Relative ACP5 protein level in 95C and A549 cells after different transfection, * p < 0.05, ** p < 0.01, compared with the NC group. LC, lung cancer; ACP5, tartrate-resistant acid phosphatase 5; NC, negative control
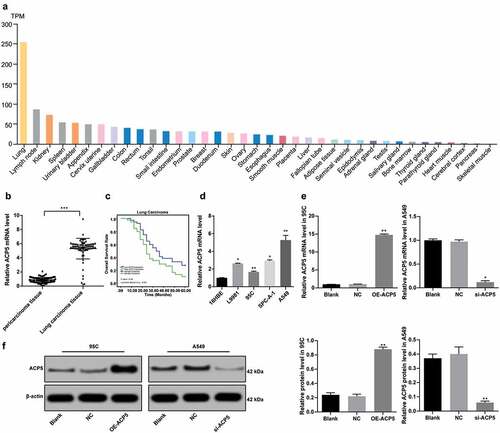
To further verify the role of ACP5 in LC progression, ACP5 expression in LC cells and tracheal epithelial cells was detected. The results of RT-qPCR showed that ACP5 expression in LC cells L9981, 95C, SPC-A-1 and A549 was noticeably higher than that in tracheal epithelial cells 16HBE (), p < 0.05). ACP5 overexpression vector was constructed and transfected into 95C cells with lower relative ACP5 expression. ACP5 si-RNA vector was constructed and transfected into A549 cells with relative highest ACP5 expression. The results of RT-qPCR and western blot analysis demonstrated that ACP5 expression was interfered, indicating successful transfection ().
3.2. ACP5 knockdown inhibits LC cell growth and induces apoptosis
Considering ACP5 expression was highly expressed in LC, and related to clinical stage, differentiation degree and lymph node metastasis of LC patients, now we turned to explore effects of ACP5 inhibition in molecular processes of LC. MTT assay detected cell viability and showed increased cell viability in 95C cells with overexpressed ACP5, while decreased cell viability in A549 cells with inhibited ACP5 expression (), p < 0.05). Flow cytometry detected cell proliferation and showed increased cell proliferation rate in 95C cells with overexpressed ACP5, while decreased cell proliferation in A549 cells with inhibited ACP5 expression (), p < 0.05). PCNA only exists in mitotic cells, and PCNA expression represents the proliferation ability of cells [Citation21]. PCNA immunofluorescence assay showed increased number of positive cells in 95C cells with overexpressed ACP5, while decreased number of positive cells in A549 cells with inhibited ACP5 expression (), p < 0.05). These results suggested ACP5 knockdown could promote LC cell proliferation.
Figure 2. ACP5 knockdown inhibits LC cell growth and induces apoptosis. a. Relative cell viability after transfection with overexpressed ACP5 or si-ACP51 measured by MTT assay; b. Relative cell proliferation after transfection with overexpressed ACP5 or si-ACP51 measured by flow cytometry; c. Representative images and statistical chart of relative PCNA positive cells after transfection with overexpressed ACP5 or si-ACP51 measured by PCNA immunofluorescence assay; d. Representative images and statistical chart of relative cell apoptosis after transfection with overexpressed ACP5 or si-ACP51 measured by flow cytometry; e. Representative images of PI staining after transfection with overexpressed ACP5 or si-ACP51; f. Protein expression of Bax, PUMA and Cleaved PARP after transfection with overexpressed ACP5 or si-ACP51 detected by western blot analysis; g. Representative images and statistical chart of cell cycle distribution after transfection with overexpressed ACP5 or si-ACP51 measured by flow cytometry. * p < 0.05, compared with the NC group. LC, lung cancer; ACP5, tartrate-resistant acid phosphatase 5; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide; PCNA, proliferating cell nuclear antigen; PI, propidium iodide; Bax, B-cell lymphoma-2 (Bcl-2) associated X; PARP, poly(ADP-ribose) polymerase; NC, negative control
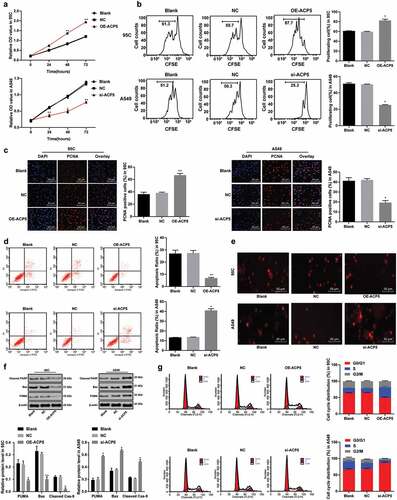
Flow cytometry measured cell cycle distribution and apoptosis, and revealed that apoptotic cells decreased notably in 95C cells with overexpressed ACP5, but no cell cycle arrest appeared; while apoptotic cells increased substantially in A549 cells with inhibited ACP5 expression, with more cells arrested in G2/M phase (, )). PI staining indicated pyknotic nucleus decreased in 95C cells with overexpressed ACP5, while damaged nuclei increased in A549 cells with inhibited ACP5 expression ()). Western blot analysis measured apoptotic-related proteins Bax, PUMA and Cleaved poly(ADP-ribose) polymerase (PARP) showing decreased levels of apoptotic-related proteins in 95C cells with overexpressed ACP5, but increased in A549 cells with inhibited ACP5 expression (), all p < 0.05). Thus, we concluded that ACP5 knockdown could induce LC cell apoptosis.
3.3. RUNX1 can specifically bind to ACP5 promoter
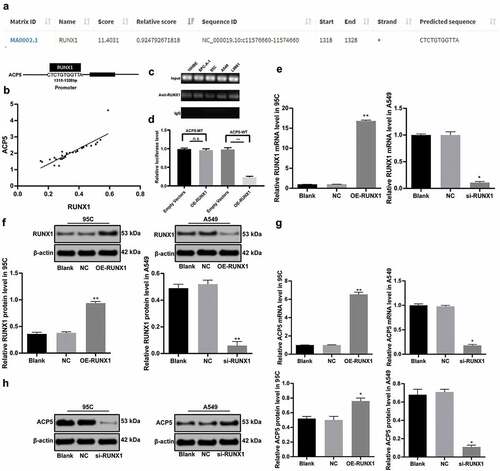
Transcription factors, as an important regulator of gene expression, can promote or inhibit gene expression. A former study has shown that abnormal expression of transcription factors can promote cancer occurrence [Citation22]. Therefore, we screened the transcription factor RUNX1 through an online prediction website JASPAR (http://jaspar.genereg.net/), which could act on ACP5 promoter and promote its transcription ()), and the action site is shown in ). Subsequently, the correlation between mRNA expression of ACP5 and RUNX1 was analyzed. The results showed a positive correlation ()). The results of ChIP assay and dual luciferase reporter gene assay demonstrated that RUNX1 could bind specifically to ACP5 promoter (). RUNX1 overexpression vector was constructed and transfected into 95C cells, and RUNX1 si-RNA was constructed and transfected into A549 cells. The results of RT-qPCR and western blot analysis showed RUNX1 expression was interfered, indicating successful transfection (). Moreover, RT-qPCR and western blot analysis detected the ACP5 expression after RUNX1 silencing, and the results suggested silencing RUNX1 resulted in decreased ACP5 expression (, p < 0.05).
Figure 3. RUNX1 can bind specifically to ACP5 promoter. a. Prediction correlation between ACP5 and RUNX1 promoter, and the action site; b. correlation between mRNA expression of ACP5 and RUNX1; c and d. RUNX1 could bind specifically to ACP5 promoter measured by ChIP assay and dual luciferase reporter gene assay; e. Relative RUNX1 mRNA expression after transfection with overexpression vector and si-RNA vectors detected by RT-qPCR; f. Relative RUNX1 protein level after transfection with overexpression vector and si-RNA vectors determined by western blot analysis; g. Relative ACP5 mRNA expression after transfection with overexpression vector and si-RNA vectors detected by RT-qPCR; h. Relative ACP5 protein level after transfection with overexpression vector and si-RNA vectors determined by western blot analysis. * p < 0.05, ** p < 0.01, compared with the NC group. LC, lung cancer; ACP5, tartrate-resistant acid phosphatase 5; RUNX1, runt-related transcription factor 1; ChIP, chromatin immunoprecipitation; si-RNA, small interfering RNA; NC, negative control
3.4. RUNX1 inhibition suppresses LC cell growth and induces apoptosis
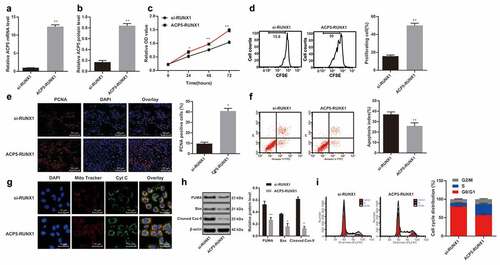
After knowing the effects of ACP5 on LC cell growth and RUNX1 could bind specifically to ACP5 promoter, now the focus of this study shifted to the investigation of mechanisms of RUNX1 in LC progression. MTT assay detected cell viability and showed increased cell viability in 95C cells with overexpressed RUNX1, while decreased cell viability in A549 cells with inhibited RUNX1 expression (), p < 0.05). Flow cytometry detected cell proliferation and showed increased cell proliferation rate in 95C cells with overexpressed RUNX1, while decreased cell proliferation in A549 cells with inhibited RUNX1 expression (), p < 0.05). PCNA immunofluorescence assay showed increased number of positive cells in 95C cells with overexpressed RUNX1, while decreased number of positive cells in A549 cells with inhibited RUNX1 expression (), p < 0.05). These results suggested RUNX1 knockdown could promote LC cell proliferation.
Figure 4. RUNX1 knockdown inhibits LC cell growth and induces apoptosis. a. Relative cell viability after transfection with overexpression RUNX1 or si-RUNX1 measured by MTT assay; b. Relative cell proliferation after transfection with overexpression RUNX1 or si-RUNX1 measured by flow cytometry; c. Representative images and statistical chart of relative PCNA positive cells after transfection with overexpression RUNX1 or si-RUNX1 measured by PCNA immunofluorescence assay; d. Representative images and statistical chart of relative cell apoptosis after transfection with overexpression RUNX1 or si-RUNX1 measured by flow cytometry; e. Representative images of PI staining after transfection with overexpression RUNX1 or si-RUNX1; f. Relative levels of Bax, PUMA and Cleaved PARP after transfection with overexpression RUNX1 or si-RUNX1 detected by western blot analysis; g. Relative cell cycle distribution after transfection with overexpression RUNX1 or si-RUNX1 measured by flow cytometry. * p < 0.05, ** p < 0.01, compared with the NC group. LC, lung cancer; RUNX1, runt-related transcription factor 1; ACP5, tartrate-resistant acid phosphatase 5; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide; PCNA, proliferating cell nuclear antigen; PI, propidium iodide; Bax, B-cell lymphoma-2 (Bcl-2) associated X; NC, negative control
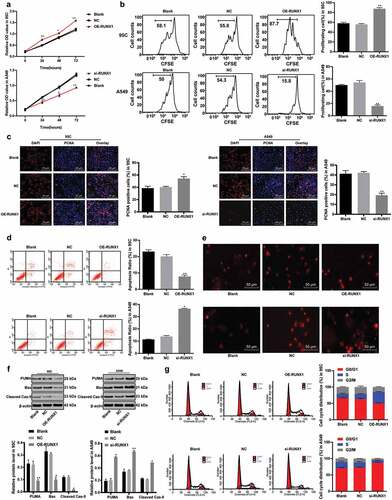
Flow cytometry measured cell cycle distribution and apoptosis and revealed that apoptotic cells decreased notably in 95C cells with overexpressed RUNX1, but no cell cycle arrest appeared; while apoptotic cells increased substantially in A549 cells with inhibited RUNX1 expression, with more cells arrested in G2/M phase (, ), all p < 0.05). PI staining indicated pyknotic nucleus decreased in 95C cells with overexpressed RUNX1, while damaged nuclei increased in A549 cells with inhibited RUNX1 expression (), p < 0.05). Western blot analysis measured apoptotic-related proteins Bax, PUMA and Cleaved PARP. The results exhibited decreased levels of apoptotic-related proteins in 95C cells with overexpressed RUNX1, but increased in A549 cells with inhibited RUNX1 expression (), p < 0.05). Thus, we concluded RUNX1 knockdown could induce LC cell apoptosis.
3.5. Overexpressed ACP5 alleviates the inhibition of LC cell activity induced by silencing RUNX1
To further verify roles of ACP5 in LC development, we constructed an overexpressed ACP5 vector and co-transfected it into A549 cells carrying si-RUNX1. The results of RT-qPCR and Western blot analysis confirmed the successful transfection (). After overexpression of ACP5 and silencing RUNX1, LC cell viability and proliferation rate increased significantly (), cell apoptosis decreased notably (), and cell cycle arrest was effectively alleviated ()). In a word, overexpressed ACP5 could relieve the inhibition of LC cell activity induced by silencing RUNX1.
Figure 5. Overexpressed ACP5 relieves the inhibition of LC cell activity induced by silencing RUNX1. a. Relative ACP5 mRNA expression in A549 cells carrying si-RUNX1 with/without overexpressed ACP5 detected by RT-qPCR; b. A. Relative ACP5 protein level in A549 cells carrying si-RUNX1 with/without overexpressed ACP5 detected by western blot analysis; c. Relative cell viability in A549 cells carrying si-RUNX1 with/without overexpressed ACP5 measured by MTT assay; d. Relative cell proliferation in A549 cells carrying si-RUNX1 with/without overexpressed ACP5 measured by flow cytometry; e. Representative images and statistical chart of relative PCNA positive cells in A549 cells carrying si-RUNX1 with/without overexpressed ACP5 measured by PCNA immunofluorescence assay; f. Representative images and statistical chart of apoptosis in A549 cells carrying si-RUNX1 with/without overexpressed ACP5 measured by flow cytometry; g. Representative images of Cytochrome C staining in A549 cells carrying si-RUNX1 with/without overexpressed ACP5; h. Relative levels of Bax, PUMA and Cleaved caspase-9 in A549 cells carrying si-RUNX1 with/without overexpressed ACP5 detected by western blot analysis; i. Relative cell cycle distribution in A549 cells carrying si-RUNX1 with/without overexpressed ACP5 measured by flow cytometry. * p < 0.05, ** p < 0.01, compared with the si-RUNX1 group. N = e3. LC, lung cancer; ACP5, tartrate-resistant acid phosphatase 5; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide; PCNA, proliferating cell nuclear antigen; PI, propidium iodide; Bax, B-cell lymphoma-2 (Bcl-2) associated X1; RUNX1, runt-related transcription factor 1
3.6. ACP5 knockdown inhibits LC tumor growth in vivo
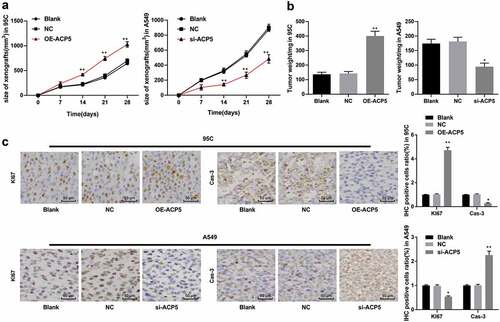
Given that ACP5 knockdown could inhibit LC cell growth and induce apoptosis in vitro, we conducted in vivo experiment for further validation. By measuring the growth of xenograft tumors in mice, the results showed overexpression of ACP5 in 95C cells substantially increased the volume growth and weight of xenograft tumors, which were opposite to those in A549 cells with inhibited ACP5 () p < 0.05). Immunohistochemistry was used to detect the expression of Ki67 and Caspase-3 in xenograft tumors. The results revealed that overexpression of ACP5 in 95C cells greatly increased the positive rate of Ki67 and decreased the positive rate of Caspase-3, which were reversed by ACP5 knockdown in A549 cells (), p < 0.05). As expected, ACP5 knockdown could inhibit LC tumor growth in vivo.
Figure 6. ACP5 knockdown inhibits LC tumor growth in vivo. a. The curve of xenograft tumor volume in mice with overexpressed ACP5 or inhibited ACP5; b. Relative xenograft tumor weight in mice with overexpressed ACP5 or inhibited ACP5; c. Representative images and statistical chart of positive rate of Ki67 and PARP in mice with overexpressed ACP5 or inhibited ACP5 determined by immunohistochemistry. * p < 0.05, ** p < 0.01, compared with the NC group. LC, lung cancer; ACP5, tartrate-resistant acid phosphatase 5; NC, negative control
3.7. ACP5 knockdown inhibits LC progression by blocking the ERK/MAPK pathway
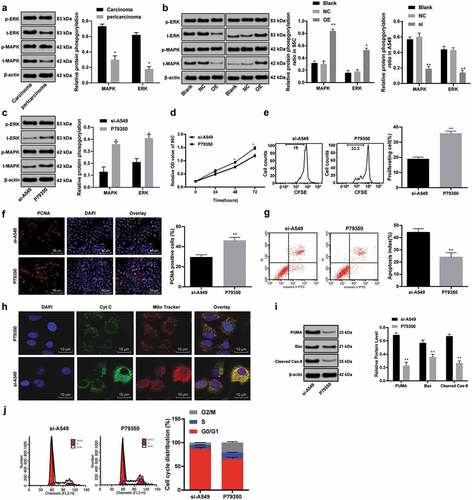
It has been researched that ERK/MAPK is highly expressed in NSCLC [Citation23], so we used Western blot analysis to detect the protein levels and phosphorylation levels of ERK/MAPK in LC tissues and paracancerous tissues. The results indicated the phosphorylation levels of ERK/MAPK in LC tissues were evidently higher than those in paracancerous tissues ()). Moreover, overexpression of ACP5 in 95C cells increased the phosphorylation levels of ERK/MAPK, while ACP5 inhibition in A549 cells decreased the phosphorylation levels of ERK/MAPK ()). Subsequently, the specific activator P79350 [Citation24] of MAPK were added into A549 cells carrying si-ACP5. Western blot analysis exhibited increased phosphorylation levels of MAPK and ERK ()), enhanced cell activity and proliferation (), reduced apoptosis (), and effectively alleviated cell cycle arrest (), all p < 0.05) in A549 cells with silencing ACP5 and P79350. In summary, ACP5 knockdown could inhibit LC progression by blocking the ERK/MAPK pathway.
Figure 7. ACP5 knockdown inhibits LC progression by blocking the ERK/MAPK pathway. a. Relative protein levels and phosphorylation levels of ERK/MAPK in LC tissues and paracancerous tissues detected by western blot analysis; b. Relative phosphorylation levels of ERK/MAPK in 95C and A549 cells with overexpressed ACP5 or si-ACP5 detected by western blot analysis; c. Relative phosphorylation levels of ERK/MAPK in A549 cells with si-ACP5 and MAPK activator P79350 detected by western blot analysis; d. Relative cell viability in A549 cells with si-ACP5 and MAPK activator P79350 measured by MTT assay; e. Relative cell proliferation in A549 cells with si-ACP5 and MAPK activator P79350 measured by flow cytometry; f. Representative images and statistical chart of PCNA positive cells in A549 cells with si-ACP5 and MAPK activator P79350 measured by PCNA immunofluorescence assay; g. Representative images and statistical chart of cell apoptosis in A549 cells with si-ACP5 and MAPK activator P79350 measured by flow cytometry; h. Representative images of Cytochrome C staining in A549 cells with si-ACP5 and MAPK activator P79350; i. Relative levels of Bax, PUMA and Cleaved caspase-9 in A549 cells with si-ACP5 and MAPK activator P79350 measured by flow cytometry; j. Relative cell cycle distribution in A549 cells with si-ACP5 and MAPK activator P79350 measured by flow cytometry. * p < 0.05, ** p < 0.01, compared with the si-ACP5 group. LC, lung cancer; ACP5, tartrate-resistant acid phosphatase 5; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide; PCNA, proliferating cell nuclear antigen; PI, propidium iodide; Bax, B-cell lymphoma-2 (Bcl-2) associated X; RUNX1, Runt-related transcription factor 1; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase
4. Discussion
Due to lacking recognized markers in the early stage, the 5-year survival rate of LC patients at regional stages was only 24.3%, and dropping dramatically to 3.6% for 56% patients at distant stages [Citation25]. Although novel molecular-targeted treatment methods have brought satisfactory efficacy in some cancer types, no targeted therapeutic approach can be applied to most LC patients so far [Citation26]. It was previously stated that serum ACP5b activity is a potentially useful marker in diagnosing and monitoring bone metastasis, a gravely debilitating complication of NSCLC, and provide greater diagnostic sensitivity for early detection and monitoring treatment response [Citation27]. In this study, we assumed there may be roles of ACP5 and RUNX1 in LC cell growth with the involvement of the ERK/MAPK pathway. Consequently, our findings revealed silenced RUNX1 could inhibit LC progression by downregulating the ERK/MAPK pathway and binding to ACP5.
We firstly found ACP5 was highly expressed in LC patients and cells, and ACP5 expression in cancer tissues was positively correlated with higher tumor stage, lower differentiation, and lymph node metastasis. As reported recently in a research mainly made by Gao YL, ACP5 was upregulated in lung adenocarcinoma tissues, and its overexpression showed a strong link with lymph node metastasis, differentiation and bad prognosis [Citation12], which further supported the association of ACP5 expression with LC progression. Nai-Shun Yao et al. reported that in NSCLC patients, alveolar macrophages could be activated by cancer itself, leading to increased serum ACP5b activity [Citation27]. Overexpression of ACP5 was also observed in other tumors. For example, ACP5 was also upregulated in hepatocellular carcinoma tissues and accompanied with larger tumor size, microvascular invasion, poor differentiation and higher tumor-node-metastasis (TNM) stage [Citation28], which further verified the carcinogenic role of ACP5 in solid tumors. Besides, our results revealed ACP5 knockdown could inhibit LC cell growth and induce apoptosis, presenting by increased levels of Bax, PUMA and Cleaved PARP. Bax can trigger the activation of caspase through mitochondrial dysfunction, leading to the release of pro-apoptotic promoters into the cytoplasm, resulting in cell death finally [Citation29]. PUMA plays an essential role in activating apoptosis through both p53 dependent and independent pathways [Citation30]. Previously, exogenous PUMA transfection was reported to inhibit NSCLC cell proliferation and promote apoptosis [Citation31]. Consistent observation of ACP5 knockdown inhibiting cell proliferation and invasion was found in melanoma cells, demonstrating the protective roles of ACP5 knockdown in malignant tumors [Citation32].
Furthermore, we revealed that the function of ACP5 in LC progression was achieved via the regulation of RUNX1 and the ERK/MAPK pathway. Previous evidences suggested RUNX transcription factors play pivotal roles in various human tumors [Citation33]. Our results demonstrated RUNX1 could bind specifically to ACP5 promoter, and silencing RUNX1 resulted in decreased ACP5 expression. Similarly, RUNX1 expression was markedly upregulated in colorectal cancer tissues [Citation34] and genetic variation in RUNX1 was associated with high risk for colon and rectal cancers [Citation35]. Besides, we demonstrated RUNX1 knockdown could inhibit LC cell growth and induce apoptosis, presenting by increased levels of Bax, PUMA and Cleaved PARP. RUNX1 knockdown increased levels of pro-apoptotic proteins, Bax and Cleaved caspase-3, and inhibited proliferation in colorectal cancer [Citation36]. Xianghui Wang et al. confirmed RUNX1 was highly expressed in LC and RUNX1 knockout increased LC cell chemosensitivity and inhibited cell invasion [Citation37]. Additionally, ACP5 knockdown inhibits LC progression by blocking the ERK/MAPK pathway. Levels of ERK and MAPK were higher in NSCLC tissues were associated with lymph node metastasis and TNM stage of NSCLC patients [Citation23]. MAPK and ERK1/2 can regulate apoptosis by phosphorylating tumor suppressor p53 in LC cells [Citation38]. Inhibition of NF-κB activity in osteoclasts precursors further inhibited expression of NF-κB downstream target ACP5 [Citation39]. Of particular note, p53 eliminated DNA-damaged cells by inducing irreversible apoptosis by activating its downstream genes Bax and PUMA [Citation40], in which RUNX1 was required to stimulate p53 through complex formation and acetylation [Citation41]. Interestingly, p53 mutation could activate NF-κB pathway, repress levels of pro-apoptotic proteins Bax, and then enhance chemo resistance and tumorigenesis of LC [Citation42].
Together, these results offer insights into the mechanism of ACP5, RUNX1 and the ERK/MAPK pathway in LC development, whereby RUNX1 silencing could prevent LC progression by downregulating the ERK/MAPK pathway and binding to ACP5. Analysis of abovementioned results may better understand the background for future clinical investigation and application in LC molecular treatment. Although our findings offer therapeutic implication in LC treatment, the experiment results and effective application into clinical practice need further validation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311.
- Yoon SM, Shaikh T, Hallman M. Therapeutic management options for stage III non-small cell lung cancer. World J Clin Oncol. 2017;8:1–20.
- Bach PB, Mirkin JN, Oliver TK, et al. Benefits and harms of CT screening for lung cancer: a systematic review. JAMA. 2012;307:2418–2429.
- Reck M, Popat S, Reinmuth N, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii27–39.
- Mao Y, Yang D, He J, et al. Epidemiology of Lung Cancer. Surg Oncol Clin N Am. 2016;25:439–445.
- Lemjabbar-Alaoui H, Hassan OU, Yang YW, et al. Lung cancer: biology and treatment options. Biochim Biophys Acta. 2015;1856:189–210.
- Vlachogeorgos GS, Manali ED, Blana E, et al. Placental isoform glutathione S-transferase and P-glycoprotein expression in advanced nonsmall cell lung cancer: association with response to treatment and survival. Cancer. 2008;114:519–526.
- Haley KJ, Lasky-Su J, Manoli SE, et al. RUNX transcription factors: association with pediatric asthma and modulated by maternal smoking. Am J Physiol Lung Cell Mol Physiol. 2011;301:L693–701.
- Khan M, Cortes J, Kadia T, et al. Clinical outcomes and Co-occurring mutations in patients with RUNX1-mutated acute myeloid leukemia. Int J Mol Sci. 2017;18:1618.
- Tang X, Sun L, Jin X, et al. Runt-related transcription factor 1 regulates LPS-induced acute lung injury via NF-kappaB signaling. Am J Respir Cell Mol Biol. 2017;57:174–183.
- Halling Linder C, Ek-Rylander B, Krumpel M, et al. Bone alkaline phosphatase and tartrate-resistant acid phosphatase: potential Co-regulators of bone mineralization. Calcif Tissue Int. 2017;101:92–101.
- Gao YL, Liu MR, Yang SX, et al. Prognostic significance of ACP5 expression in patients with lung adenocarcinoma. Clin Respir J. 2018;12:1100–1105.
- Jiang M, Zhou LY, Xu N, et al. Hydroxysafflor yellow A inhibited lipopolysaccharide-induced non-small cell lung cancer cell proliferation, migration, and invasion by suppressing the PI3K/AKT/mTOR and ERK/MAPK signaling pathways. Thorac Cancer. 2019;10:1319–1333.
- Boucherat O, Landry-Truchon K, Aoidi R, et al. Lung development requires an active ERK/MAPK pathway in the lung mesenchyme. Dev Dyn. 2017;246:72–82.
- Wang L, Brugge JS, Janes KA. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc Natl Acad Sci U S A. 2011;108:E803–12.
- Zhang S, Chen S, Liu A, et al. Inhibition of BDNF production by MPP(+) through up-regulation of miR-210-3p contributes to dopaminergic neuron damage in MPTP model. Neurosci Lett. 2018;675:133–139.
- Xu T, Niu C, Zhang X, et al. beta-Ecdysterone protects SH-SY5Y cells against beta-amyloid-induced apoptosis via c-Jun N-terminal kinase- and Akt-associated complementary pathways. Lab Invest. 2018;98:489–499.
- Ji Y, Han Z, Shao L, et al. Evaluation of in vivo antitumor effects of low-frequency ultrasound-mediated miRNA-133a microbubble delivery in breast cancer. Cancer Med. 2016;5:2534–2543.
- Huang C, Liu W, Perry CN, et al. Autophagy and protein kinase C are required for cardioprotection by sulfaphenazole. Am J Physiol Heart Circ Physiol. 2010;298:H570–9.
- Yoo JJ, Lee JH, Lee SH, et al. Comparison of the effects of transarterial chemoembolization for advanced hepatocellular carcinoma between patients with and without extrahepatic metastases. PLoS One. 2014;9:e113926.
- Leng F, Saxena L, Hoang N, et al. Proliferating cell nuclear antigen interacts with the CRL4 ubiquitin ligase subunit CDT2 in DNA synthesis-induced degradation of CDT1. J Biol Chem. 2018;293:18879–18889.
- Wu L, Wang P. Long non-coding RNA-neighboring enhancer of FOXA2 inhibits the migration and invasion of small cell lung carcinoma cells by downregulating transforming growth factor-beta1. Oncol Lett. 2019;17:4969–4975.
- Cao Q, Mao ZD, Shi YJ, et al. MicroRNA-7 inhibits cell proliferation, migration and invasion in human non-small cell lung cancer cells by targeting FAK through ERK/MAPK signaling pathway. Oncotarget. 2016;7:77468–77481.
- Lv B, Huo F, Dang X, et al. Puerarin attenuates N-methyl-D-aspartic acid-induced apoptosis and retinal ganglion cell damage through the JNK/p38 MAPK pathway. J Glaucoma. 2016;25:e792–801.
- Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241.
- Li C, Lyu J, Meng QH. MiR-93 promotes tumorigenesis and metastasis of non-small cell lung cancer cells by activating the PI3K/Akt pathway via inhibition of LKB1/PTEN/CDKN1A. J Cancer. 2017;8:870–879.
- Yao NS, Wu YY, Janckila AJ, et al. Serum tartrate-resistant acid phosphatase 5b (TRACP5b) activity as a biomarker for bone metastasis in non-small cell lung cancer patients. Clin Chim Acta. 2011;412:181–185.
- Xia L, Huang W, Tian D, et al. ACP5, a direct transcriptional target of FoxM1, promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Oncogene. 2014;33:1395–1406.
- Lee JS, Jung WK, Jeong MH, et al. Sanguinarine induces apoptosis of HT-29 human colon cancer cells via the regulation of Bax/Bcl-2 ratio and caspase-9-dependent pathway. Int J Toxicol. 2012;31:70–77.
- Zhao L, Zhang HY, Pang YK, et al. [Knockdown of Puma protects cord blood CD34(+) cells against gamma- irradiation]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014;22:412–420.
- Liu CJ, Zhang XL, Luo DY, et al. Exogenous p53 upregulated modulator of apoptosis (PUMA) decreases growth of lung cancer A549 cells. Asian Pac J Cancer Prev. 2015;16:741–746.
- Scott KL, Nogueira C, Heffernan TP, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20:92–103.
- Kayed H, Jiang X, Keleg S, et al. Regulation and functional role of the Runt-related transcription factor-2 in pancreatic cancer. Br J Cancer. 2007;97:1106–1115.
- Kara M, Yumrutas O, Ozcan O, et al. Differential expressions of cancer-associated genes and their regulatory miRNAs in colorectal carcinoma. Gene. 2015;567:81–86.
- Slattery ML, Lundgreen A, Herrick JS, et al. Associations between genetic variation in RUNX1, RUNX2, RUNX3, MAPK1 and eIF4E and riskof colon and rectal cancer: additional support for a TGF-beta-signaling pathway. Carcinogenesis. 2011;32:318–326.
- Zhou Y, Zhang X, Zhang J, et al. LRG1 promotes proliferation and inhibits apoptosis in colorectal cancer cells via RUNX1 activation. PLoS One. 2017;12:e0175122.
- Wang X, Zhao Y, Qian H, et al. The miR-101/RUNX1 feedback regulatory loop modulates chemo-sensitivity and invasion in human lung cancer. Int J Clin Exp Med. 2015;8:15030–15042.
- Taylor CA, Zheng Q, Liu Z, et al. Role of p38 and JNK MAPK signaling pathways and tumor suppressor p53 on induction of apoptosis in response to Ad-eIF5A1 in A549 lung cancer cells. Mol Cancer. 2013;12:35.
- Tai YT, Landesman Y, Acharya C, et al. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications. Leukemia. 2014;28:155–165.
- Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442.
- Wu D, Ozaki T, Yoshihara Y, et al. Runt-related transcription factor 1 (RUNX1) stimulates tumor suppressor p53 protein in response to DNA damage through complex formation and acetylation. J Biol Chem. 2013;288:1353–1364.
- Yang L, Zhou Y, Li Y, et al. Mutations of p53 and KRAS activate NF-kappaB to promote chemoresistance and tumorigenesis via dysregulation of cell cycle and suppression of apoptosis in lung cancer cells. Cancer Lett. 2015;357:520–526.
