ABSTRACT
Small nucleolar RNA host gene 7 (SNHG7) is a newly recognized oncogenic Long non-coding RNA (lncRNA) in most human cancers. In gastric cancer, SNHG7 has been suggested to enhance cell proliferation and suppressed apoptosis through down-regulating P15 and P16 expression, but the effect of SNHG7 on gastric cancer cell migration and invasion was still unknown. In our study, we aimed to estimate the relationship between SNHG7 expression and clinical and pathological characteristics, and explore the effect of SNHG7 on gastric cancer cell migration and invasion. In our study, the levels of SNHG7 expression in gastric cancer tissues and cell lines were severally higher than in normal adjacent tissues and gastric mucosal epithelial cells. Moreover, high SNHG7 expression was positively correlated with TNM stage, depth of invasion, lymph-node metastasis and distant metastasis in gastric cancer patients. Furthermore, the multivariate Cox proportional hazard analysis further showed high SNHG7 expression was an independent poor prognostic factor for overall survival in gastric cancer patients. The studies in vitro revealed that SNHG7 directly binds to miR-34a and negatively regulates miR-34a expression, and SNHG7 enhances gastric cancer cell migration and invasion through suppressing miR-34a-Snail-EMT axis. In conclusion, SNHG7 functions as oncogenic lncRNA in gastric cancer and may be a potential therapeutic target for gastric cancer patients.
Abbreviations: lncRNA: Long non-coding RNA; SNHG7: Small nucleolar RNA host gene 7; EMT: Epithelial mesenchymal transition; TNM: Tumor-Lymph Node-Metastasis.
Introduction
Gastric cancer is the fifth common malignant disease and the second leading cause of cancer deaths worldwide, with about 1,033,701 new cases and 782,685 deaths from the disease in 2018 [Citation1]. In China, there has been a persistent and steady decline in incidence and mortality of gastric cancer patients over the last decades [Citation2]. However, gastric cancer remains the second frequently diagnosed type of tumor and the second leading cause of cancer-related death in China [Citation3]. Despite of the progress in diagnosis and treatment of gastric cancer, the clinical outcome is still unsatisfactory, with a 5-y survival of only 21.35% [Citation4]. Therefore, a deeper understanding of the mechanism under gastric cancer progression could provide valuable biomarkers and potential therapeutic targets for improving the prognosis of gastric cancer.
Long non-coding RNAs (lncRNAs) are a novel member of non-coding RNAs, which is longer than 200 nucleotides in length and lacks protein-coding capacity [Citation5,Citation6]. Small nucleolar RNA host gene 7 (SNHG7) is a newly recognized oncogenic lncRNA that is located on chromosome 9q34.3 [Citation7]. The published study showed that SNHG7 was overexpressed in gastric cancer tissues and cells, and enhanced cell proliferation and suppressed apoptosis through down-regulating P15 and P16 expression [Citation8]. The effect of SNHG7 on gastric cancer cell migration and invasion was still unknown. In our preliminary study, we found gastric cancer patients with positive lymph-node metastasis had a higher level of SNHG7 expression than those with negative lymph-node metastasis. In addition, SNHG7 has been suggested to participate in modulating tumor cell metastasis, such as lung cancer [Citation9,Citation10], breast cancer [Citation11,Citation12], hepatocellular carcinoma [Citation13,Citation14], colorectal cancer [Citation15,Citation16], pancreatic cancer [Citation17], bladder cancer [Citation18–Citation20], prostate cancer [Citation21], osteosarcoma [Citation22], nasopharyngeal carcinoma [Citation23], melanoma [Citation24], neuroblastoma [Citation25] and glioblastoma [Citation26]. Therefore, we guessed SNHG7 is involved in regulating gastric cancer metastasis. In our study, we aimed to estimate the relationship between SNHG7 expression and clinical and pathological characteristics, and explore the effect of SNHG7 on gastric cancer cell migration and invasion.
Materials and methods
Clinical samples
This study included 162 gastric cancer patients who underwent surgical operation or biopsy at The First Affiliated Hospital of Soochow University or Xuzhou Central Hospital between 2013 and 2018. Normal adjacent tissues were obtained at least 3 cm away from the primary tumors. All clinical tissues were immediately snap frozen in liquid nitrogen and then stored at −80℃. The histological diagnosis of all clinical tissues was confirmed by at least two pathologists. None of gastric cancer patients received any anti-tumor treatment prior to surgery or biopsy. The study was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University or Xuzhou Central Hospital. All patients have signed written informed consent forms prior to the experiment.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
The qRT-PCR was performed to detect the expression of SNHG7 and miR-34a in gastric cancer tissues, normal adjacent tissues, normal human gastric mucosa epithelial cells and gastric cancer cells by, respectively, using TB Green Premix Ex Taq II (Takara Biotechnology, Dalian, China) and Mir-X miRNA First Strand Synthesis Kit (Takara Biotechnology, Dalian, China) as described previously [Citation27]. The following primers were used: SNHG7 forward, 5'-TTGCTGGCGTCTCGGTTAAT-3' and reverse, 5'-GGAAGTCCATCACAGGCGAA-3'; GAPDH forward, 5'-TCCTCTGACTTCAACAGCGACAC-3' and reverse, 5'-CACCCTGTTGCTGTAGCCAAATTC-3'; miR-34a forward, 5'-TGGCAGTGTCTTAGCTGGTTG-3'; U6 forward, 5'-GCGCGTCGTGAAGCGTTC-3'. The reverse primers for miR-34a and U6 were provided in Mir-X miRNA First Strand Synthesis Kit. SNHG7 and miR-34a were, respectively, normalized to GAPDH and U6. The relative expression was calculated using the 2−ΔΔCT method.
Cell lines and cell transfection
Human gastric cancer cell lines (MKN-45, SGC-7901, N87) and human gastric mucosal epithelial cells (GES-1) were cultured at Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Rockville, MD, USA) containing 10% fetal bovine serum (FBS, Gibco, Rockville, MD, USA) at 37℃ in a humidified 5% CO2 atmosphere.
The full-length sequences of SNHG7 were inserted into pcDNA3.1 (+) and named as pc-SNHG7. The small interfering RNAs targeting SNHG7 (si-SNHG7) and miR-34a mimics and miR-34a inhibitor were synthesized by GenePharma Co., Ltd. (Shanghai, China). Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was used for cell transfection as described previously [Citation27].
Luciferase reporter assay
The potential binding sites of SNHG7 and miR-34a were predicted on starBase (http://starbase.sysu.edu.cn/). To construct luciferase reporter vectors, SNHG7 fragments containing the predicted miR-34a binding site were amplified and cloned into the pmiR-RB-REPORT luciferase reporter plasmid. Luciferase assays were performed using the Dual Luciferase Reporter assay system (Promega, Madison, WI, USA) as described previously [Citation27].
Transwell migration and invasion assays
Transwell migration and invasion assays were performed by transwell chamber inserts (24-wells, 8 μm pore size, 6.5 mm diameter; Corning, NY, USA) and matrigel matrix (1:8; 50 μl/well; BD Biosciences, Franklin Lakes, NJ, USA) as described previously [Citation27].
Western blot
The Snail, Ecadherin and Vimentin protein levels were detected by western blot according to previous study [Citation28]. The primary antibodies in this research were showed as follow: anti-Snail, anti-Ecadherin, anti-Vimentin and anti-β-actin (dilution 1:1000; Cell Signaling Technology, Danvers, MA, USA). β-actin was used as an internal control.
Statistical analysis
Statistical analyses were performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). All data were shown as mean ± standard deviation from three independent experiments. Differences between two independent groups were measured by the Student t-test. Correlations between SNHG7 expression and clinicopathological characteristics were estimated by Chi-square test or Fisher’s exact test. The correlation between SNHG7 and miR-34a in gastric cancer tissues was analyzed with Pearson's correlation test. Survival curves were generated and analyzed by the Kaplan–Meier method and log-rank test. Univariate and multivariate analysis was performed by Cox regression model. P < 0.05 was considered to indicate a statistically significant difference.
Results
The expression of SNHG7 in gastric cancer tissues and cell lines
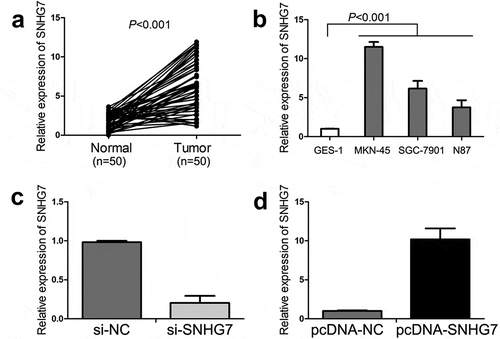
The expression of SNHG7 in gastric cancer tissues and normal adjacent tissues was measured through qRT-PCR. The result of qRT-PCR showed SNHG7 was significantly increased in gastric cancer tissues compared with normal adjacent tissues (P < 0.001, ). Meanwhile, we also detected the levels of SNHG7 expression in gastric cancer cell lines (MKN-45, SGC-7901, N87) and gastric mucosal epithelial cells (GES-1) by qRT-PCR. As shown in , the levels of SNHG7 expression in gastric cancer cell lines were higher than in gastric mucosal epithelial cells (P < 0.001). Based on the expression pattern of SNHG7 expression in gastric cancer cell lines (), MKN-45 cells were chosen for loss-of-function study and N87 cells were chosen for gain-of-function study. Subsequently, qRT-PCR was used to evaluate the transfection efficiency of si-SNHG7 and pcDNA-SNHG7. As shown in , the SNHG7 expression was markedly down-regulated in MKN-45 cells by si-SNHG7 transfection and was obviously up-regulated in N87 cells by pcDNA-SNHG7 transfection.
Figure 1. The expression of SNHG7 in gastric cancer tissues and cell lines.
The clinical value of SNHG7 in gastric cancer patients
In order to explore the correlations between SNHG7 expression and clinicopathological features in gastric cancer patients, all gastric cancer cases were divided into low SNHG7 expression group and a high SNHG7 expression group based on the median value of SNHG7 expression. We observed that high SNHG7 expression was positively correlated with TNM stage (P = 0.014, ), depth of invasion (P = 0.001, ), lymph-node metastasis (P = 0.004, ) and distant metastasis (P < 0.001, ) in gastric cancer patients. However, there was no significant correlation of SNHG7 expression with gender (P = 0.193, ), age (P > 0.748, ) and Lauren’s classification (P = 0.058, ).
Table 1. Correlations between SNHG7 expression and clinicopathological characteristics in gastric cancer patients
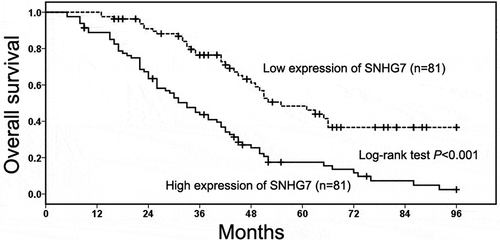
The prognostic value of SNHG7 in gastric cancer patients
The relationship between SNHG7 expression and the overall survival of gastric cancer patient was analyzed through the Kaplan–Meier method and log-rank test. We found gastric cancer patients in high SNHG7 expression group had short overall survival compared with patients in low SNHG7 expression group (P < 0.001, ). Then, the univariate Cox proportional hazard analysis was conducted to identify prognostic factor in gastric cancer patients, and the results showed TNM stage (P < 0.001, ), depth of invasion (P < 0.001, ), lymph-node metastasis (P < 0.001, ), distant metastasis (P < 0.001, ) and SNHG7 expression (P < 0.001, ) were prognostic factors. Furthermore, the multivariate Cox proportional hazard analysis further showed high SNHG7 expression was an independent poor prognostic factor for overall survival in gastric cancer patients (P = 0.032, ).
Table 2. Univariate and multivariate Cox regression analyses of overall survival in gastric cancer paitents
The relationship between SNHG7 and miR-34a in gastric cancer
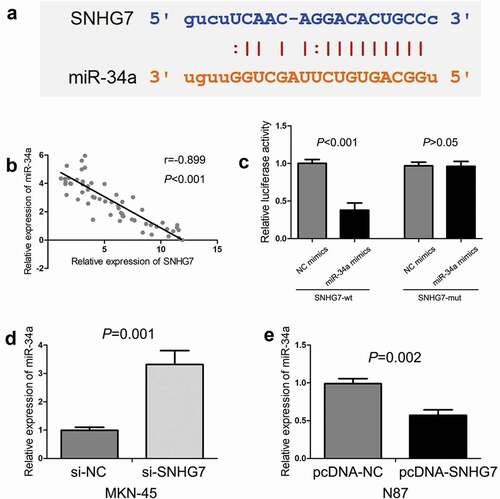
Bioinformatics online tool (starBase) was conducted to predict the potential target miRNAs for SNHG7, revealing the complementary binding sites with miR-34a (). In addition, we observed there was a negative correlation between SNHG7 expression and miR-34a expression in gastric cancer tissues (P < 0.001, ). Furthermore, we conducted luciferase reporter assay for confirming the potential binding sites between SNHG7 and miR-34a, and found miR-34a mimics could decrease the luciferase activity with SNHG7-wt (P < 0.001, ), but miR-34a mimics cannot decrease the luciferase activity with SNHG7-mut (P > 0.05, ). Then, we estimated the effect of SNHG7 on miR-34a expression in gastric cancer cells, and found down-regulation of SNHG7 elevated the level of miR-34a expression in MKN-45 cells (P < 0.001, ), and up-regulation of SNHG7 reduced the level of miR-34a expression in N87 cells (P < 0.001, ). Therefore, results indicated that SNHG7 directly binds to miR-34a, and negatively regulates miR-34a expression.
Figure 3. The relationship between SNHG7 and miR-34a in gastric cancer.
SNHG7 enhances gastric cancer cell migration and invasion through suppressing miR-34a
For exploring the influence of SNHG7 on gastric cancer cell migration and invasion, we performed a loss-of-function study in MKN-45 cells and gain-of-function study in N87 cells. The results of transwell migration and invasion assays showed down-regulation of SNHG7 obviously inhibited gastric cancer cell migration and invasion (P < 0.001, P < 0.001, ), and up-regulation of SNHG7 remarkably accelerated gastric cancer cell migration and invasion (P < 0.001, P < 0.001, ).
Figure 4. SNHG7 enhances gastric cancer cell migration and invasion through suppressing miR-34a.
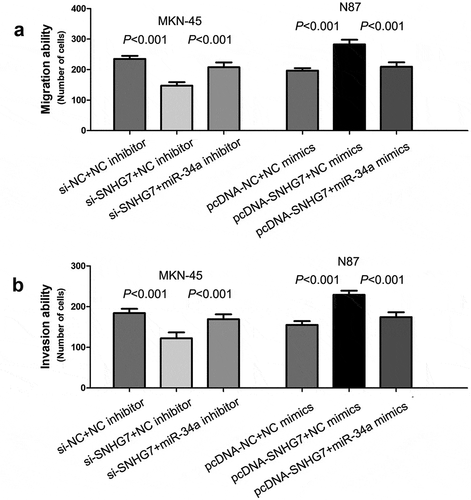
The miR-34a has been suggested to play an important role in regulating tumor cell migration and invasion [Citation29–Citation32]. Above results showed that SNHG7 directly binds to miR-34a, and negatively regulates miR-34a expression. Therefore, we performed rescue experiments, and found that co-transfection of si-SNHG7 and miR-34a inhibitor rescued the inhibition of si-SNHG7 on TNBC cell migration and invasion (P < 0.001, P < 0.001, ), and co-transfection of pcDNA-SNHG7 and miR-34a mimics rescued the acceleration of pcDNA-SNHG7 on TNBC cell migration and invasion (P < 0.001, P < 0.001, ).
SNHG7 regulates miR-34a-Snail-EMT axis in gastric cancer cells
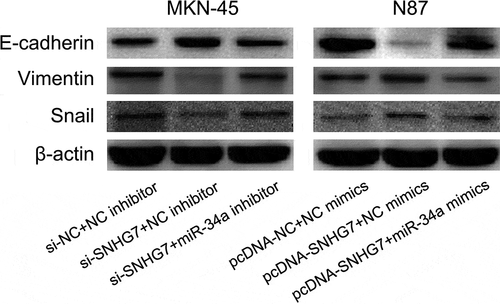
The miR-34a-Snail-epithelial mesenchymal transition (EMT) axis has been showed to play a critical role in cancer progression and metastasis [Citation33–Citation36]. Meanwhile, we found SNHG7 enhances gastric cancer cell migration and invasion through suppressing miR-34a. Thus, we supposed SNHG7 regulates the miR-34a-Snail-EMT axis in gastric cancer cells. Firstly, we conducted western blot to estimate the effect of DLX6‑AS1 on Snail, E-cadherin and Vimentin protein expression. We found down-regulation of SNHG7 elevated E-cadherin protein expression and reduced Snail and Vimentin protein expression in MKN-45 cells, and up-regulation of SNHG7 decreased E-cadherin protein expression and increased Snail and Vimentin protein expression in N87 cells (). Subsequently, we performed a rescue experiment for exploring whether SNHG7 mediates miR-34a to regulate Snail, E-cadherin and Vimentin protein expression in gastric cancer cells. We observed miR-34a inhibitor abated the impact of si-SNHG7 on modulating Snail, E-cadherin, and Vimentin protein expression, and miR-34a mimics rescue the impact of pcDNA-SNHG7 on regulating Snail, E-cadherin, and Vimentin protein expression in gastric cancer cells ().
Discussion
SNHG7 is a newly recognized lncRNA, which has been suggested to be involved in a great deal of pathophysiological processes such as ischemic stroke [Citation37], cardiac hypertrophy [Citation38], diabetic retinopathy [Citation39], recurrent spontaneous abortion [Citation40] and fracture repair [Citation41]. Furthermore, there was more evidence indicating that SNHG7 plays a critical role in human tumorigenesis and progression. SNHG7 has been suggested to be overexpressed in most types of human cancer including lung cancer [Citation9,Citation10], breast cancer [Citation11,Citation12], hepatocellular carcinoma [Citation14], colorectal cancer [Citation15,Citation16], pancreatic cancer [Citation17], bladder cancer [Citation18–Citation20], prostate cancer [Citation21,Citation42], osteosarcoma [Citation22,Citation43], nasopharyngeal carcinoma [Citation23], melanoma [Citation24], neuroblastoma [Citation25], glioblastoma [Citation26], esophageal cancer [Citation44] and hypopharyngeal cancer [Citation45]. In gastric cancer, we found that the levels of SNHG7 expression in gastric cancer tissues and cell lines were severally higher than in normal adjacent tissues and gastric mucosal epithelial cells. Wang M.W. et al. also found SNHG7 expression was overexpressed in gastric cancer tissues and cell lines compared with tumor-adjacent normal tissues and normal gastric cell line, respectively [Citation8]. However, the correlations between SNHG7 expression and clinicopathological features in gastric cancer patients were still unknown. In our study, we further explored the clinical value of SNHG7 in gastric cancer patients through analyzing the correlations between SNHG7 expression and clinicopathological features, and observed that high SNHG7 expression was positively correlated with TNM stage, depth of invasion, lymph-node metastasis and distant metastasis. Similarly, She Kelin et al. reported SNHG7 overexpression was associated with high TNM stage and positive lymph-node metastases in patients with non-small cell lung cancer [Citation10]. Moreover, Shan Yujia et al. and Li Yang et al. conformably showed high SNHG7 expression was significantly associated with present lymphatic metastasis and distant metastasis in colorectal cancer patients [Citation15,Citation16]. In pancreatic cancer patients, Cheng Dongfeng et al. indicated that SNHG7 overexpression was correlated with advanced TNM stage, positive lymph-node metastasis, large tumor size and unfavorable tumor differentiation [Citation17]. Besides, the expression of SNHG7 was found to be related to bladder patient’s tumor size, lymphatic metastasis and clinical stage [Citation18–Citation20]. Similar clinical value of SNHG7 was also observed in breast cancer [Citation11], prostate cancer [Citation42], osteosarcoma [Citation22,Citation43], neuroblastoma [Citation25], esophageal cancer [Citation44] and hypopharyngeal cancer [Citation45]. Generally, high SNHG7 expression is correlated with clinical progression, implying that SNHG7 has a close relationship to patient’s survival time.
The prognostic significance of SNHG7 has been investigated in several types of human cancer. High SNHG7 expression has favorable or unfavorable prognostic significance depending on cancer types. In chromophobe renal cell carcinoma patients, He Hai-Tao et al. suggested SNHG7 expression levels were positively correlated with overall survival time [Citation46]. There were more studies suggesting SNHG7 overexpression predicted unfavorable prognosis, such as breast cancer [Citation11], colorectal cancer [Citation15,Citation16], pancreatic cancer [Citation17], bladder cancer [Citation19], osteosarcoma [Citation22,Citation43], neuroblastoma [Citation25], glioblastoma [Citation26], prostate cancer [Citation42] and hypopharyngeal cancer [Citation45]. Li Yang et al. performed multivariate Cox regression analyses, and observed SNHG7 overexpression was an independent unfavorable factor of the prognosis in colorectal cancer patients [Citation16]. Moreover, Chi Renjie et al. suggested high SNHG7 expression acted as an independent poor prognostic predictor for overall survival in neuroblastoma patients [Citation25]. In hypopharyngeal cancer patients, Ping Wu et al. also found high SNHG7 expression served as an independent unfavorable prognostic factor for overall survival time [Citation45]. The clinical value of SNHG7 in gastric cancer patients was still unknown. We found gastric cancer patients in high SNHG7 expression group had short overall survival compared with patients in low SNHG7 expression group. Furthermore, multivariate Cox proportional hazard analysis showed high SNHG7 expression was an independent poor prognostic factor for overall survival in gastric cancer patients. Generally, SNHG7 is a valuable prognostic biomarker in many human cancers.
SNHG7 has been suggested to enhance cell proliferation and suppress apoptosis through down-regulating P15 and P16 expression [Citation8]. The effect of SNHG7 on gastric cancer cell migration and invasion was still unknown. Our clinical analysis showed high SNHG7 expression was positively correlated with the depth of invasion, lymph-node metastasis and distant metastasis. Thus, we further explored the influence of SNHG7 on gastric cancer cell migration and invasion, and we found the down-regulation of SNHG7 inhibited gastric cancer cell migration and invasion, and up-regulation of SNHG7 accelerated gastric cancer cell migration and invasion. For investigating the molecular mechanism of SNHG7, we found SNHG7 has a close relationship with miR-34a through bioinformatics analysis and correlation analysis. The miR-34a has been showed to function as tumor suppressive microRNA to regulate cell migration and invasion through modulating Snail-EMT axis [Citation33–Citation36]. In addition, Sun Xianfu et al. reported that SNHG7 targeted miR-34a and promoted EMT initiation to enhance breast cancer cell proliferation and invasion [Citation12]. Moreover, Deng Yiqi et al. indicated that SNHG7 promote tumor growth and EMT process through suppressing miR-34a [Citation22]. In our study, we found SNHG7 enhanced gastric cancer cell migration and invasion through suppressing miR-34a-Snail-EMT axis. Generally, SNHG7 functions as oncogenic lncRNA in gastric cancer.
In conclusion, high SNHG7 expression is associated with clinical progression and prognosis. SNHG7 enhances gastric cancer cell migration and invasion through suppressing miR-34a-Snail-EMT axis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Zheng RS, Sun KX, Zhang SW, et al. Report of cancer epidemiology in China, 2015. Zhonghua Zhong Liu Za Zhi. 2019;41(1):19–28.
- Chen JG, Chen HZ, Zhu J, et al. Cancer survival in patients from a hospital-based cancer registry, China. J Cancer. 2018;9(5):851–860.
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981.
- Gugnoni M, Ciarrocchi A. Long noncoding RNA and Epithelial Mesenchymal transition in cancer. Int J Mol Sci. 2019;20(8):E1924.
- Chaudhry MA. Expression pattern of small nucleolar RNA host genes and long non-coding RNA in X-rays-treated lymphoblastoid cells. Int J Mol Sci. 2013;14(5):9099–9110.
- Wang MW, Liu J, Liu Q, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21(20):4613–4622.
- She K, Huang J, Zhou H, et al. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36(5):2673–2680.
- She K, Yan H, Huang J, et al. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell Prolif. 2018;51(1):e12406.
- Luo X, Song Y, Tang L, et al. LncRNA SNHG7 promotes development of breast cancer by regulating microRNA-186. Eur Rev Med Pharmacol Sci. 2018;22(22):7788–7797.
- Sun X, Huang T, Liu Z, et al. LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR-34a through EMT initiation and the Notch-1 pathway. Eur J Pharmacol. 2019;856:172407.
- Cui H, Zhang Y, Zhang Q, et al. A comprehensive genome-wide analysis of long noncoding RNA expression profile in hepatocellular carcinoma. Cancer Med. 2017;6(12):2932–2941.
- Sun BZ, Ji DG, Feng ZX, et al. Long noncoding RNA SNHG7 represses the expression of RBM5 to strengthen metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(13):5699–5704.
- Shan Y, Ma J, Pan Y, et al. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9(7):722.
- Li Y, Zeng C, Hu J, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol. 2018;11(1):89.
- Cheng D, Fan J, Ma Y, et al. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019;9:28.
- Zhong X, Long Z, Wu S, et al. LncRNA-SNHG7 regulates proliferation, apoptosis and invasion of bladder cancer cells assurance guidelines. J Buon. 2018;23(3):776–781.
- Chen Y, Peng Y, Xu Z, et al. Knockdown of lncRNA SNHG7 inhibited cell proliferation and migration in bladder cancer through activating Wnt/beta-catenin pathway. Pathol Res Pract. 2019;215(2):302–307.
- Xu C, Zhou J, Wang Y, et al. Inhibition of malignant human bladder cancer phenotypes through the down-regulation of the long non-coding RNA SNHG7. J Cancer. 2019;10(2):539–546.
- Han Y, Hu H, Zhou J. Knockdown of LncRNA SNHG7 inhibited epithelial-mesenchymal transition in prostate cancer though miR-324-3p/WNT2B axis in vitro. Pathol Res Pract. 2019;215(10):152537.
- Deng Y, Zhao F, Zhang Z, et al. Long noncoding RNA SNHG7 promotes the tumor growth and Epithelial-to-Mesenchymal transition via regulation of miR-34a signals in Osteosarcoma. Cancer Biother Radiopharm. 2018;33(9):365–372.
- Wang L, Xu T, Cui X, et al. Downregulation of lncRNA SNHG7 inhibits proliferation and invasion of nasopharyngeal carcinoma cells through repressing ROCK1. Eur Rev Med Pharmacol Sci. 2019;23(14):6186–6193.
- Zhang C, Zhu B, Li XB, et al. Long non-coding RNA SNHG7 promotes migration and invasion of melanoma via upregulating SOX4. Eur Rev Med Pharmacol Sci. 2019;23(11):4828–4834.
- Chi R, Chen X, Liu M, et al. Role of SNHG7-miR-653-5p-STAT2 feedback loop in regulating neuroblastoma progression. J Cell Physiol. 2019;234(8):13403–13412.
- Ren J, Yang Y, Xue J, et al. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496(2):712–718.
- Zhang Y, Li Y, Han L, et al. SUMO1P3 is associated clinical progression and facilitates cell migration and invasion through regulating miR-136 in non-small cell lung cancer. Biomed Pharmacother. 2019;113:108686.
- Yuan Y, Yangmei Z, Rongrong S, et al. Sotrastaurin attenuates the stemness of gastric cancer cells by targeting PKCdelta. Biomed Pharmacother. 2019;117:109165.
- Hu Y, Pu Q, Cui B, et al. MicroRNA-34a inhibits tumor invasion and metastasis in gastric cancer by targeting Tgif2. Int J Clin Exp Pathol. 2015;8(8):8921–8928.
- Zhou Y, Ding BZ, Lin YP, et al. MiR-34a, as a suppressor, enhance the susceptibility of gastric cancer cell to luteolin by directly targeting HK1. Gene. 2018;644:56–65.
- Peng Y, Guo JJ, Liu YM, et al. MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci Rep. 2014;34(3):e00112.
- Wei B, Huang QY, Huang SR, et al. MicroRNA34a attenuates the proliferation, invasion and metastasis of gastric cancer cells via downregulation of MET. Mol Med Rep. 2015;12(4):5255–5261.
- Wang Y, Wu Z, Hu L. The regulatory effects of metformin on the [SNAIL/miR-34]:[ZEB/miR-200] system in the epithelial-mesenchymal transition(EMT) for colorectal cancer(CRC). Eur J Pharmacol. 2018;834:45–53.
- Tang Y, Tang Y, Cheng YS. miR-34a inhibits pancreatic cancer progression through Snail1-mediated epithelial-mesenchymal transition and the Notch signaling pathway. Sci Rep. 2017;7:38232.
- Hahn S, Jackstadt R, Siemens H, et al. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. Embo J. 2013;32(23):3079–3095.
- Siemens H, Jackstadt R, Hunten S, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10(24):4256–4271.
- Zheng Y, Sun S, Yu M, et al. Identification of potential hub-lncRNAs in ischemic stroke based on subpathway-LNCE method. J Cell Biochem. 2019;120(8):12832–12842.
- Jing L, Li S, Wang J, et al. Long non-coding RNA small nucleolar RNA host gene 7 facilitates cardiac hypertrophy via stabilization of SDA1 domain containing 1 mRNA. J Cell Biochem. 2019;120(9):15089–15097.
- Ke N, Pi LH, Liu Q, et al. Long noncoding RNA SNHG7 inhibits high glucose-induced human retinal endothelial cells angiogenesis by regulating miR-543/SIRT1 axis. Biochem Biophys Res Commun. 2019;514(2):503–509.
- Xiang H, Yan H, Sun B, et al. Decreased expression of long non-coding RNA SNHG7 cause recurrent spontaneous abortion through suppression proliferation and invasion of trophoblast cells via miR-34a. Am J Transl Res. 2019;11(1):463–472.
- Chen Z, Liu Z, Shen L, et al. Long non-coding RNA SNHG7 promotes the fracture repair through negative modulation of miR-9. Am J Transl Res. 2019;11(2):974–982.
- Qi H, Wen B, Wu Q, et al. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother. 2018;102:326–332.
- Zhang GD, Gai PZ, Liao GY, et al. LncRNA SNHG7 participates in osteosarcoma progression by down-regulating p53 via binding to DNMT1. Eur Rev Med Pharmacol Sci. 2019;23(9):3602–3610.
- Xu LJ, Yu XJ, Wei B, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur Rev Med Pharmacol Sci. 2018;22(9):2653–2661.
- Wu P, Tang Y, Fang X, et al. Metformin suppresses Hypopharyngeal cancer growth by Epigenetically silencing long non-coding RNA SNHG7 in FaDu cells. Front Pharmacol. 2019;10:143.
- He HT, Xu M, Kuang Y, et al. Biomarker and competing endogenous RNA potential of tumor-specific long noncoding RNA in chromophobe renal cell carcinoma. Onco Targets Ther. 2016;9:6399–6406.