ABSTRACT
Objective: Long noncoding RNAs (lncRNAs) have already been proposed to function in Parkinson’s disease (PD). However, the role of lncRNA BACE1-AS in PD has never been discussed. This study aims to examine the mechanism of BACE1-AS on oxidative stress injury of dopaminergic neurons in PD rats.
Methods: Rat models of PD were established through the injection of 6-hydroxydopamine. The rotation of rats was induced by intraperitoneal injection of apomorphine, and number of rotations per minute was detected. The levels of malondialdehyde (MDA), superoxide dismutase (SOD), and glutathione peroxidase (GSH-Px), glutamic acid (Glu), dopamine (DA), tyrosine hydroxylase (TH), α-synuclein and inducible nitric oxide synthase (iNOS) in the substantia nigra of rats in each group were detected. Apoptosis and pathological changes in the substantia nigra were also observed. BACE1-AS, miR-34b-5p, BACE1, Bax and Bcl-2 expression in the substantia nigra were detected. The binding of BACE1-AS and miR-34b-5p and the targeting relationship of miR-34b-5p and BACE1 were further determined.
Results: Downregulated BACE1-AS reduced iNOS, α-synuclein and Glu levels and elevated DA and TH levels in the substantia nigra of PD rats. Downregulated BACE1-AS repressed apoptosis and oxidative stress injury in the substantia nigra neurons of PD rats. BACE1-AS specifically bound to miR-34b-5p. BACE1 was a direct target gene of miR-34b-5p.
Conclusion: Collectively, our study reveals that downregulation of lncRNA BACE1-AS inhibits iNOS activation in the substantial nigra and improve oxidative stress injury in PD rats by upregulating miR-34b-5p and downregulating BACE1.
Introduction
Parkinson’s disease (PD) refers to a neurodegenerative disorder manifested by motor and nonmotor symptoms, such as tremor, rigidity, bradykinesia, hyposmia and psychosis [Citation1,Citation2]. Many risk factors are accountable for the occurrence of PD, including dairy products, pesticides, cancer, methamphetamine, traumatic brain injury, blood cholesterol and hypertension [Citation3]. Due to the complexity of PD, clinical challenges remain, including incapability to definitively diagnose this disease at the earliest stages and difficulties of symptom management at later stages, and no treatments have been found to inhibit the neurodegenerative process [Citation4]. This emphasizes the urgent need to develop some promising therapies for PD treatment.
Long noncoding RNAs (lncRNAs) refer to non-protein-coding RNA molecules which are over 200 nt in length [Citation5]. LncRNAs form big networks of ribonucleoprotein (RNP) complexes through many chromatin regulators, and at the same time, target these enzymatic activities to proper locations in the genome, which makes lncRNAs modular scaffolds to specify higher order organization in chromatin states and RNP complexes [Citation6]. An early report has shown that lncRNA BACE1-AS is overexpressed in Alzheimer disease (AD) along with the RNA-binding protein HuD that is capable of binding both β-Site amyloid precursor protein cleaving enzyme 1 (BACE1) and BACE1-AS transcripts and enhancing their stability [Citation7]. Evidence has suggested that lncRNA HOTAIR elevates Leucine-rich repeat kinase 2 expression to boost-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced PD [Citation8]. Similar study by Cao et al. has revealed that lncRNA SNHG1 contributes to neuroinflammation in PD by modulating microRNA-7 (miR-7)/NLRP3 pathway [Citation9]. MiRNAs refer to short noncoding RNAs that target specific genes posttranscriptionally to control gene expression, and are discovered to regulate fundamental cellular processes [Citation10]. There has been a literature showing that miR-34b-5p, a member of the miR-34 family, regulates hippocampal astrocyte apoptosis in flurothyl-induced recurrent seizures [Citation11]. Results from a previous study have also demonstrated that repression of miR-34b and miR-34 c elevates α-synuclein expression in PD, which further promotes PDpathogenesis [Citation12]. BACE1 is the main neuronal β-secretase for amyloid-β generation and is degraded in lysosomes [Citation13]. It has been indicated that suppression of BACE1 activity could negatively work on AD progression by controlling amyloid β peptide production [Citation14]. However, there has been no study focusing on the role of lncRNA BACE1-AS in PD. Thus, this study intends to discuss the function of the BACE1-AS/miR-34b-5p/BACE1 axis in PD rats.
Materials and methods
Ethics statement
Animals were treated humanely in compliance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of Institute for Brain Sciences Research, School of Life Sciences, Henan University.
Experimental animals
Eighty clean and healthy male Sprague Dawley rats aging 6–8 wk old and weighing 200 ± 12 g were purchased from Hunan SJA Laboratory Animal Co., Ltd. (Hunan, China). The rats were raised at a temperature of 24 ± 1°C with 55 ± 20% humidity and received normal light. Rats were kept in a terrarium before and after modeling and had free access to sufficient food and water. The relevant equipment was employed after autoclaving for the prevention of cross-contamination.
Modeling and grouping
Rats were anesthetized through intraperitoneal injection of 3% pentobarbital sodium (1.5 mL/kg) and fixed on stereotaxic apparatus. According to the stereotaxic map of the rat brain (George Paxinos & Charles Watson), the skull was drilled (location: 5.0 mm before bregma, 2.1 mm left of midline, 7.7 mm under endocranium), and injected with 2 μL lentivirus (packed and synthesized by Shanghai Genechem Co., Ltd. Shanghai, China) carrying target gene or an equal amount of normal saline with a microsyringe on the left substantia nigra. Eighty rats were divided into eight groups: normal group (injected with normal saline), 6-hydroxydopamine (6-OHDA) group (injected with normal saline, and 6-OHDA was used to induce PD model), overexpression-negative control (oe-NC) group (injected with BACE1-AS overexpression lentivirus NC 1 wk before modeling), oe-BACE1-AS group (injected with BACE1-AS overexpression lentivirus 1 wk before modeling), sh-NC group (injected with BACE1-AS silencing lentivirus NC 1 wk before modeling), sh-BACE1-AS group (injected with BACE1-AS silencing lentivirus 1 wk before modeling), and sh-BACE1-AS + inhibitors NC group (injected with BACE1-AS silencing and miR-34b-5p inhibitor lentivirus NC 1 wk before modeling), sh-BACE1-AS + miR-34b-5p inhibitors group (injected with BACE1-AS silencing and miR-34b-5p inhibitor lentivirus 1 wk before modeling).
During modeling, the rats in each group were fixed by stereotaxic apparatus and the skull was drilled (location: 4.2 mm before bregma, 0.9 mm left of midline, 7.8 mm under endocranium and 5.3 mm after bregma, 1.8 mm right of midline, 7.8 mm under endocranium). PD models were established by injecting 2 μL 6-OHDA (4 μg/mL) into the ventral midbrain and its black dense parts of rats, and the normal group was injected with the same volume of normal saline (1 μL/minute; the needle was detained for 10 min after injection, and slowly withdrawn at 1 mm/min). After the operation, the operation area was lightly wiped with hydrogen peroxide, and the borehole was filled with gelatin sponge, sutured, and disinfected. The rats were removed from the stereotaxic apparatus, caged separately, and given enough room to keep warm, feed, and drink water. Postoperative rats were given an intraperitoneal injection of penicillin 50,000 U/d for 7 d to prevent infection. Rats were weighed once every 3 d and observed for health record.
Behavioral testing
Rotation test was induced by intraperitoneal injection of apomorphine (Sigma-Aldrich, St. Louis, MO, USA) at 0.5 mg/kg 1 wk after modeling. The number of rat rotations to the right within 30 min was recorded, and the rotational speed ≥7 r/min was regarded as successful modeling of PD rats [Citation15]. Rotation test was performed once a week for 3 consecutive weeks and the average was taken.
Substantia nigra tissues extraction and processing
After behavioral test, the rats in each group were euthanatized by decapitation before thoracotomy. The heart was firstly perfused with normal saline with 1% heparin for blood removal, and then the heart was perfused with 50 mL mixture of 4% paraformaldehyde (PFA) and 2.5% glutaraldehyde of 4°C (first fast and then slow). Next, a craniotomy was rapidly applied for brain tissue extraction and isolation. The fresh brain tissue in the substantia nigra was quickly taken out from rat brain placed on an ice tray with ophthalmic forceps, frozen in liquid nitrogen in a cryotube, and stored at −80°C for further experiments.
Hematoxylin-eosin (HE) staining
The substantia nigra sections were dewaxed with xylene for 20 min, dehydrated with gradient concentration of alcohol (100%, 95%, 80%, 75%) for 1 min, rinsed with distilled water for 2 min, and stained with hematoxylin for 10 min. Then the sections were differentiated with hydrochloric acid ethanol for 30 s, soaked in warm water at 50°C for 5 min before 30-s eosin staining. Subsequently, 70% and 90% alcohol were adopted for dehydration for 10 min each, followed by xylene permeabilization, sealing with neutral gum, and observation of tissue morphology at high magnification.
Transmission electron microscope (TEM) observation
The spare substantia nigra tissues were double-fixed with 2.5% glutaraldehyde and 1% citric acid, dehydrated, and embedded with Epon812 resin. Then, the tissues were cut into semi-thin sections and stained with toluidine blue. Next, ultra-thin sections were prepared after trimming the tissue masses before double staining of uranyl acetate and lead citrate. Finally, the sections were observed by JME-2000EX TEM (JEOL, Ltd., Tokyo, Japan).
Immunohistochemistry
The substantia nigra tissues stored in the refrigerator were fixed in 4% PFA and placed in 20% sucrose solution at 4°C overnight. Then, the tissues were sliced and dewaxed before 10-min incubation with 2 drops of 3% H2O2 at room temperature. Next, primary antibody hydroxylase (TH) (1:750, Abcam, Cambridge, MA, USA) and inducible nitric oxide synthase (iNOS) (1:100, Abcam, Cambridge, MA, USA) were added for 2-h incubation, along with horseradish peroxidase-labeled immunoglobulin G(IgG) for 30 min at room temperature. Subsequently, the sections were stained with 50 μL diaminobenzidine (DAB) at 25°C for 5 min, counterstained with hematoxylin, and differentiated with 0.1% hydrochloric acid, followed by alcohol dehydration, permeabilization with xylene, and sealing with neutral gum. After dried, the sections were observed at high magnification for positive staining area rate calculation.
Terminal deoxyribonucleotidyl transferase (TDT)-mediated dUTP-digoxigenin nick end labeling (TUNEL) staining
The prepared substantia nigra sections were washed in xylene twice (5 min each time), dehydrated with gradient alcohol (100%, 95%, 80%, 75%, 3 min each), and treated with 20 μg/mL DNase-free proteinase K for 15–30 min. Then, the sections were supplemented with 50 μL TUNEL solution (Boster Biological Technology Co., Ltd., Wuhan, China) at 37°C for 60 min, stained with 50 μL DAB at 25°C for 10 min, counterstained with hematoxylin, and dehydrated with gradient alcohol. After permeabilization with xylene and sealing with neutral gum, the sections were observed under a light microscope for apoptosis rate calculation.
Oxidative stress level detection
Enzyme-linked immunosorbent assay (ELISA) was adopted to detect changes in oxidative stress indices in rat substantia nigra. Substantia nigra tissue was supplemented with an appropriate amount of normal saline, and stirred with tissue grinder under ice bath until all tissues were liquefied before 10-min centrifugation at 8000 r/min at 4°C and supernatant collection. The contents of malondialdehyde (MDA), superoxide dismutase (SOD), and glutathione peroxidase (GSH-Px) were measured according to the instructions of the kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
High-performance liquid chromatography (HPLC) detection of glutamic acid (Glu) content
The mobile phase methanol was used as a solvent to gradually dilute the Glu mother liquor, and an appropriate amount of the standard solution was placed in the injection bottle before adding the derivatizing reagent and the sodium tetraborate buffer. A standard curve was established using the Agilent 1200 HPLC (Agilent Technologies, 1200 series, Palo Alto, CA, USA) with the HPLC peak area and corresponding concentration for different concentrations of Glu standards. The spare substantia nigra tissue was supplemented with normal saline at 1:9, and homogenized in an ice bath. Next, the homogenate was centrifuged at 3000 r/min at low temperatures for 15 min, and 0.5 mL supernatant was taken and supplemented with 1 mol/L HClO4 before centrifuged at 10,000 r/min for 10 min. Subsequently, 0.5 mL supernatant was taken again, supplemented with 1 mL 2 mol/L K2 CO3, and centrifuged at 10,000 r/min for 20 min. Then, 30 μL supernatant was taken and mixed with an equal volume of o-phthalaldehyde derivative. After a 2-min reaction at room temperature, sample injection was performed (detection was the same as that of the standard). The peak area of Glu in each sample was obtained in HPLC, and the corresponding concentration was calculated using the established standard curve equation. The final Glu content in the sample was obtained by conversion based on the dilution factor.
HPLC with electrochemical detection (HPLC-EC) of dopamine (DA) content
Sample pretreatment: 80 mg substantia nigra was taken and supplemented with 490 μL 0.1 mol/L perchloric acid buffer (containing 0.1 mmol/L disodium ethylenediamine tetraacetic acid [EDTA-Na2]) and 10 μL internal standard solution (0.02 g/L dihydroxybenzoic acid) before ultrasonic homogenization in an ice bath and high-speed centrifugation (15,000 r/min) for 30 min at 4°C. Then, the supernatant was continuously centrifuged at 15,000 r/min for 10 min, followed by a collection of 10 μL supernatant for sample injection detection.
Mobile phase: sodium acetate (40 mmol/L) citric acid (30 mmol/L) buffer containing 0.2 mmol/L EDTA-Na2, 0.4 mmol/L sodium octyl sulfonate (B8) and 180 mL/L methanol was applied after PH value adjustment to 4.1 and 15-min ultrasonic degassing by vacuum filtration (the flow rate of mobile phase: 0.5 mL/min; the reference electrode: Ag/AgCl; the voltage of electrochemical detector: +0.8 V).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Tissue total RNA was extracted by one-step method of Trizol (Invitrogen, Carlsbad, CA, USA), and high-grade RNA was confirmed by ultraviolet spectroscopy technology and formaldehyde gel electrophoresis. Then, 1 μg RNA was taken and reverse-transcribed into cDNA with reverse transcription kits (Promega Corporation, Madison, WI, USA). The PCR primers were designed and synthesized by Beijing ComWin Biotech Co. Ltd (Beijing, China) () with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal reference of BACE1-AS, iNOS and α-synuclein, and miR-34b-5p with U6 as the internal reference. The threshold was selected manually at the lowest point of parallel rise of each logarithmic amplification curve to obtain the Ct value (Threshold cycle) in each reaction tube. The data were analyzed by 2−ΔΔCt method (2−ΔΔCt represented the multiple proportion of target genes of the experimental group and the control group) [Citation16]. The experiment was repeated three times and the data were averaged.
Table 1. Primer sequence
Western blot analysis
Total protein of tissues was extracted, and the protein concentration was measured based on the instructions of bicinchoninic acid kit (Boster Biological Technology Co., Ltd., Wuhan, China). The extracted proteins were added to the loading buffer and boiled for10 min at 95°C. Each well was loaded with 30 μg sample, and the proteins were then separated by 10% polyacrylamide gel (Boster Biological Technology Co., Ltd., Wuhan, China) through electrophoresis with the electrophoresis voltage changed from 80 v to 120 v. Subsequently, the proteins were transferred to polyvinylidene difluoride membrane through 45–70 min wet transfer with a voltage of 100 mv before 1-h 5% bovine serum albumin (BSA) sealing. Next, primary antibodies TH (1:1000), iNOS (1:1000), α-synuclein (1:1000), BACE1 (1:1000), GAPDH (1:5000) (Abcam, Cambridge, MA, USA), Bax and Bcl-2 (1:1000, Santa Cruz Biotechnology, Santa Cruz, CA., USA) were added for overnight stay at 4°C, followed by adding corresponding secondary antibody (Shanghai Miaotong Biotechnology Co., Ltd., Shanghai, China) for 1-h incubation at room temperature and development with chemiluminescent reagents. GAPDH was used as an internal reference, andGel Doc EZ Imager (Bio-rad, California, USA) for development. The gray value of target bands was analyzed with Image J software (National Institutes of Health, Bethesda, Maryland, USA). The experiment was repeated three times and the data were averaged.
Dual-luciferase reporter gene assay
The binding sites of lncRNA BACE1-AS and miR-34b-5p were predicted and analyzed at the bioinformatics website (https://cm.jefferson.edu/rna22/Precomputed/), and then verified by dual-luciferase reporter gene assay. The synthetic BACE1-AS 3ʹ-untranslated regions (3ʹ-UTR) gene fragment were introduced into the pMIR-reporter (Beijing Huayueyang Biotechnology Co., Ltd., Beijing, China) via endonuclease sites (Bamh1 and Ecor1). The complementary sequence mutation site of the seed sequence was designed on the wild type (WT) of BACE1-AS, and the target fragment was inserted into the pMIR-reporter plasmid by T4 DNA ligase after restriction endonuclease digestion. The luciferase reporter plasmids (WT and mutant type [MUT]) with correct sequence were separately co-transfected with mimics NC and miR-34b-5p mimics into 293 T cells (Shanghai North Connaught Biotechnology Co., Ltd., Shanghai, China). Forty-eight hours later, the cells were lysed for luciferase activity measurement with luciferase assay kit (BioVision, San Francisco, CA, USA) and Glomax20/20 luminometer (Promega, Madison, Wisconsin, USA). The experiment was repeated three times.
The targeting relationship of miR-34b-5p and BACE1 and the binding site of miR-34b-5p and BACE1 3ʹUTR were predicted with a bioinformatics software (https://cm.jefferson.edu/rna22/Precomputed/). The BACE1 3ʹUTR promoter sequence containing the miR-34b-5p binding site was synthesized, and a BACE1 3ʹUTR WT plasmid (BACE1-WT) was constructed. The point mutation kits (Takara Bio Inc., Otsu, Shiga, Japan) were employed to mutate the binding site of miR-34b-5p on BACE1, thereby BACE1 3ʹUTR MUT plasmid (BACE1-MUT) was established. Then, 293 T cells in the logarithmical growth phase were inoculated in 96-well plates and transfected with the mixtures of BACE1-WT and BACE1-MUT plasmids with mimics NC and miR-34b-5p mimics plasmids, respectively, by Lipofectamine 2000 at a cell density of approximately 70%. After 48 h, cells were lysed for luciferase activity detection with a luciferase assay kit. Each experiment was repeated three times.
RNA-pull down assay
The cells were separately transfected with biotin-labeledmiR-34b-5p WT plasmid and MUT plasmid (50 nM each). After 48 h, the cells were incubated with specific cell lysate (Ambion, Austin, Texas, USA) for 10 min before collection of 50 mL cell lysate sample. Next, the lysis buffer was incubated with M-280 streptavidin magnetic beads (Sigma, St. Louis, MO, USA) pre-coated with RNase-free BSA and yeast tRNA (Sigma, St. Louis, MO, USA) at 4°C for 3 h before cold lysate washes, low salt buffer wash, and a high salt buffer wash. Then, an antagonistic miR-34b-5p probe was set up as an NC. BACE1-AS expression was detected by RT-qPCR after total RNA extraction by Trizol.
Statistical analysis
The data were analyzed by SPSS 21.0 (IBM Corp., Armonk, NY, USA) statistical software. Kolmogorov–Smirnov test indicated normal distribution of the data. The results were expressed as mean ± standard deviation. The comparison between two groups was analyzed with t-test while that among multiple groups with one-way analysis of variance (ANOVA). The pairwise comparison after ANOVA analysis was performed by the least significant difference t-test (LSD-t). The difference was statistically significant at P < 0.05.
Results
Downregulated BACE1-AS reduces MDA and Glu contents and elevates SOD, GSH-Px, and DA contents in the substantia nigra tissues of PD rats
One week after modeling, rotation of rats was introduced by intraperitoneal injection of apomorphine. The number of rotations per minute increased in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, yet declined in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05; ).
Figure 1. Downregulated BACE1-AS reduces MDA and Glu contents and elevates SOD, GSH-Px, and DA contents in the substantia nigra of PD rats. A. Number of rotations per minute of rats in each group; B. MDA content in the substantia nigra of rats in each group; C. SOD and GSH-Px contents in the substantia nigra of rats in each group; D. Glu content in the substantia nigra of rats in each group; E. DA content in the substantia nigra of rats in each group; the data in the figure were all measurement data expressed as mean ± standard deviation; a, P < 0.05 vs the normal group; b, P < 0.05 vs the oe-NC group; c, P < 0.05 vs the sh-NC group; d, P < 0.05 vs the sh-BACE1-AS+inhibitors NC group
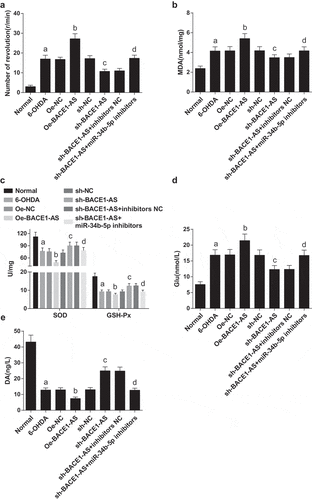
MDA, SOD and GSH-Px levels in the substantia nigra tissues of rats were detected by ELISA kits. MDA content ascended and SOD and GSH-Px contents diminished in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, while MDA content descended and SOD and GSH-Px contents grew in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05; -C).
The Glu concentration in the substantia nigra of rats was determined by HPLC. The Glu concentration in the substantia nigra of rats rose in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, yet dropped in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05;).
The DA concentration in the substantia nigra of rats was determined by HPLC-EC. The DA concentration in the substantia nigra of rats fell in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, yet grew in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05; ).
Downregulated BACE1-AS diminishes α-synuclein and iNOS level and increases TH level in the substantia nigra of PD rats
TH level in each group was detected by immunohistochemistry. It could be observed that there were brown TH-positive cells with a high concentration and normal morphology in the normal group; the 6-OHDA group, the oe-NC group, the sh-NC group and the sh-BACE1-AS+ miR-34b-5p group had less brown TH-positive cells; the oe-BACE1-AS group had seldom brown-TH positive cells; some brown TH-positive cells could be discovered in thesh-BACE1-AS group and the sh-BACE1-AS + inhibitors NC group, which were less than the normal group. It was found that the number of TH-positive cells in the substantia nigra of rats decreased in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, while the same parameter grew in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05; -B).
Figure 2. Downregulated BACE1-AS diminishes α-synuclein and iNOS level and increases TH level in the substantia nigra of PD rats. A. Detection of TH level in the substantia nigra of rats in each group by immunohistochemistry; B. Quantitative results of TH level of rats in each group; C. Detection of TH protein expression in the substantia nigra of rats in each group by Western blot analysis; D. Detection of iNOS level in the substantia nigra of rats in each group by immunohistochemistry. E. Quantitative results of iNOS level of rats in each group; F. Detection of iNOS mRNA expression in the substantia nigra of rats in each group by RT-qPCR; G: Detection of iNOS protein expression in the substantia nigra of rats in each group by Western blot analysis; H: Detection of α-synuclein mRNA expression in the substantia nigra of rats in each group by RT-qPCR; I: Detection of α-synuclein protein expression in the substantia nigra of rats in each group by Western blot analysis; the data in the figure were all measurement data expressed as mean ± standard deviation; a, P < 0.05 vs the normal group; b, P < 0.05 vs the oe-NC group; c, P < 0.05 vs the sh-NC group; d, P < 0.05 vs the sh-BACE1-AS+inhibitors NC group
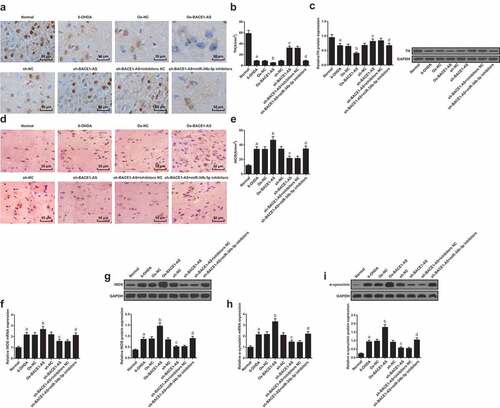
TH protein expression in the substantia nigra of rats in each group was detected by Western blot analysis. The results suggested that TH protein expression in the substantia nigra of rats in the 6-OHDA group descended in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, yet rose in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05; ).
Immunohistochemistry was adopted to detect iNOS levels in rats. Several iNOS-positive cells could be found in the normal group while there were a certain amount of iNOS-positive cells in the 6-OHDA group, the oe-NC group, the sh-NC group, and the sh-BACE1-AS + miR-34b-5p inhibitors group; the oe-BACE1-AS group had lots of iNOS-positive cells, which were more than the 6-OHDA group; the sh-BACE1-AS group and the sh-BACE1-AS+inhibitors NC group had a small number of iNOS-positive cells. The number of iNOS-positive cells in the substantia nigra of rats ascended in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, yet descended in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05; -E).
RT-qPCR and Western blot analysis were adopted to determine mRNA and protein expression of iNOS and α-synuclein in the substantia nigra of rats in each group. The results indicated that mRNA and protein expression of iNOS and α-synuclein in the substantia nigra of rats increased in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, yet decreased in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05; -I).
Downregulated BACE1-AS represses substantia nigra neuronal injury and apoptosis in PD rats
HE staining was used for observation of the substantia nigra neurons of rats in each group. The results revealed that compared with the normal group, the 6-OHDA, oe-NC, sh-NC, and sh-BACE1-AS+miR-34b-5p inhibitors groups showed obviously reduced volume of substantia nigra neurons, slightly changed nuclei, and intracellular substances centralized to edge; in the oe-BACE1-AS group, the substantia nigra neurons contracted into a round shape, and the intracellular substances were centralized obviously; the sh-BACE1-AS group and sh-BACE1-AS + inhibitorsNC group had relatively normal cell volume and less centralization of intracellular substances ().
Figure 3. Downregulated BACE1-AS represses substantia nigra neuronal injury and apoptosis in PD rats. A. Detection of the pathological changes of substantia nigra neurons of rats in each group by HE staining; B. Observation of ultrastructure of substantia nigra neurons of rats in each group by TEM; C. Detection of the apoptosis of substantia nigra neurons of rats in each group by TUNEL staining; D. Quantitative results of the apoptosis of substantia nigra neurons of rats; E. Bax and Bcl-2 protein expression in the substantia nigra of rats in each group; the data in the figure were all measurement data expressed as mean ± standard deviation; a, P < 0.05 vs the normal group; b, P < 0.05 vs the oe-NC group; c, P < 0.05 vs the sh-NC group; d, P < 0.05 vs the sh-BACE1-AS+inhibitors NC group
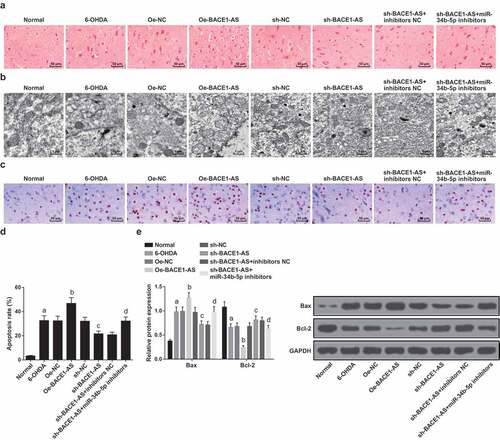
The structure of substantia nigra neurons of rats in each group was observed by TEM. The results suggested normal cell structure in the normal group: oval cell body, clear nucleolus, evenly distributed nuclear chromatin, and the rough endoplasmic reticulum, ribosome and a small number of lysosome, and Golgi complex in the cytoplasm; in the oe-BACE1-AS group, shrunken cell bodies were seen, along with segmentally fused membranes, turbid chromatin, dilated rough endoplasmic reticula, increased lysosomes, generally swollen and ruptured mitochondria and degenerated vacuoles; in the 6-OHDA, oe-NC, sh-NC, and sh-BACE1-AS + miR-34b-5p inhibitors groups, contracted cell bodies were observed, together with relatively turbid chromatin, some dilated rough endoplasmic reticula, increased lysosomes, and some swollen mitochondria; in the sh-BACE1-AS group and sh-BACE1-AS + inhibitors NC group, elliptical cells were witnessed, along with slightly smaller cell bodies, obvious nuclear membranes, slightly turbid chromatin, rough surface, slightly dilated endoplasmic reticula, and many lysosomes, individual ruptured mitochondrial membranes, and intact Golgi bodies ().
TUNEL staining was adopted to detect the apoptosis of substantia nigra neurons of rats in each group. The dark brown positive cells were apoptotic cells. The apoptotic rate of the substantia nigra of rats grew in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, yet diminished in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05; -D).
Western blot analysis was adopted to detect the expression of pro-apoptotic protein Bax and anti-apoptotic protein Bcl-2 in rat substantia nigra neurons. Bax expression rose and Bcl-2 expression fell in the substantia nigra of rats in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, while Bax expression dropped and Bcl-2 expression grew in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05;-F).
BACE1-AS specifically binds to miR-34b-5p
BACE1-AS and miR-34b-5p expression in the substantia nigra of rats in each group were detected by RT-qPCR. BACE1-AS expression ascended and miR-34b-5p descended in the nigral cells in the 6-OHDA group versus the normal group and in the oe-BACE1-AS group versus the oe-NC group, while BACE1-AS expression decreased and miR-34b-5p expression increased in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05). In the sh-BACE1-AS + miR-34b-5p inhibitors group, BACE1-AS expression was nearly the same (P > 0.05), while miR-34b-5p expression dropped (P < 0.05) in relation to the sh-BACE1-AS + inhibitors NC group ().
Figure 4. BACE1-AS specifically binds to miR-34b-5p. A. Detection of BACE1-AS and miR-34b-5p expression in the substantia nigra of rats in each group by RT-qPCR; B. Prediction of binding site of BACE1-AS and miR-34b-5p at bioinformatics site; C. Verification of the regulatory relationship between BACE1-AS and miR-34b-5p by dual-luciferase reporter gene assay. D. Verification of the binding site between BACE1-AS and miR-34b-5p by RNA-pull down assay; the data in the figure were all measurement data expressed as mean ± standard deviation; a, P < 0.05 vs the normal group; b, P < 0.05 vs the oe-NC group; c, P < 0.05 vs the sh-NC group; d, P < 0.05 vs the sh-BACE1-AS+inhibitors NC group
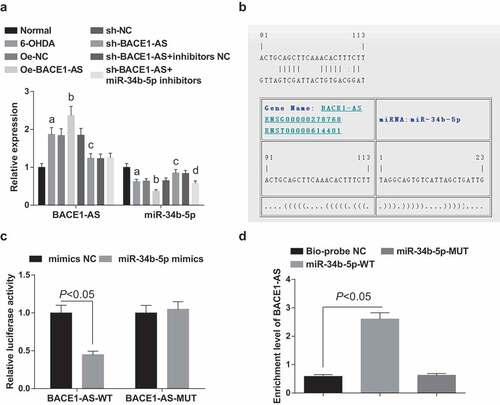
A specific binding site between the BACE1-AS gene sequence and the miR-34b-5p gene sequence was predicted by the online analysis software (). Further verification by dual-luciferase reporter gene assay indicated that the luciferase activity in the BACE1-AS-WT + miR-34b-5p mimics group descended greatly (P < 0.05), while that in the BACE1-AS-MUT +miR-34b-5p mimics group showed almost no difference (P > 0.05) in relation to the mimicsNC group, indicating that miR-34b-5p mimics might specifically bind to BACE1-AS (). RNA-pull down assay showed that BACE1-AS enrichment level in the Bio-miR-34b-5p-WT group rose (P < 0.05), while no big change in BACE1-AS enrichment level in the Bio-miR-34b-5p-MUT group was witnessed (P > 0.05) versus the Bio-probe NC group ().
BACE1 is a direct target gene of miR-34b-5p
BACE1 mRNA and protein expression in the substantia nigra of rats in each group was detected by RT-qPCR and Western blot analysis. It was found that BACE1 mRNA and protein expression in the substantia nigra of rats ascended in the 6-OHDA group versus the normal group, in the oe-BACE1-AS group versus the oe-NC group, and in the sh-BACE1-AS + miR-34b-5p inhibitors group versus the sh-BACE1-AS + inhibitors NC group, yet declined in the sh-BACE1-AS group versus the sh-NC group (all P < 0.05; -B).
Figure 5. BACE1 is a direct target gene of miR-34b-5p. A. Detection of BACE1 mRNA expression in the substantia nigra of rats in each group by RT-qPCR; B. Detection of BACE1 protein expression in the substantia nigra of rats in each group by Western blot analysis; C. Prediction of the binding site of miR-34b-5p and BACE1 at bioinformatics site; D. Verification of the regulatory relationship between miR-34b-5p and BACE1 by dual-luciferase reporter gene assay; the data in the figure were all measurement data expressed as mean ± standard deviation; a, P < 0.05 vs the normal group; b, P < 0.05 vs the oe-NC group; c, P < 0.05 vs the sh-NC group; d, P < 0.05 vs the sh-BACE1-AS+inhibitors NC group
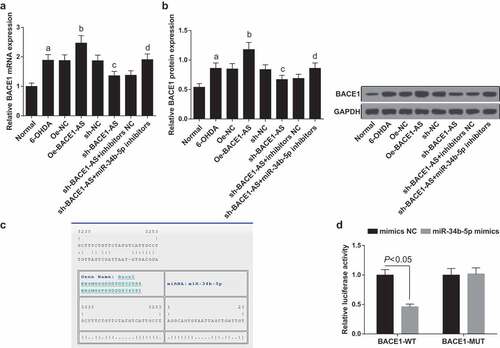
The bioinformatics software predicted a targeting site between miR-34b-5p and BACE1 (). Luciferase activity assay revealed that the relative luciferase activity of 293 T cells co-transfected with BACE1-WT and miR-34b-5p mimics reduced substantially (P < 0.05), while that of 293 T cells co-transfected with BACE1-MUT and miR-34b-5p-mimics showed no change (P > 0.05; ), suggesting that BACE1 is a direct target gene of miR-34b-5p.
Discussion
PD is a progressive neurodegenerative disorder which affects 2% of the people over 65 y old [Citation17]. The management of PD is complicated and entails the treatment of both motor and nonmotor symptoms of the disease in early and later stages [Citation18]. Lately, there has been a report suggesting that lncRNAs are abnormally expressed in PD and correlate with its pathology [Citation19]. Nevertheless, the mechanism of lncRNA BACE1-AS on PD progression remains to be discussed. Therefore, this study is conducted to examine how lncRNA BACE1-AS affects the development of PD in rats by regulating miR-34b-5p and BACE1. Collectively, our study reveals that silencing of lncRNA BACE1-AS inhibits iNOS activation in the substantial nigra to improve dopamine-dependent oxidative stress injury in PD rats by upregulating miR-34b-5p and downregulating BACE1.
Initially, we demonstrated that lncRNA BACE1-AS specifically bound to miR-34b-5p, and BACE1 was a direct target gene of miR-34b-5p. At the same time, we found that lncRNA BACE1-AS is overexpressed in PD, and knockdown of BACE1-AS elevated miR-34b-5p expression and constrained BACE1 expression. In line with our results, an early report has shown that BACE1 is a direct target gene of miR‑135b [Citation20]. A similar conclusion was reached by Gong G et al. that miR-15b directly target BACE1 mRNA 3ʹ-UTR to affect its expression [Citation21]. These basic findings are also consistent with previous research suggesting that transfection of miR-124 mimics or inhibitors leads to decrease or rise in BACE1 expression, respectively, [Citation22]. The target relation between miR-34b-5p and BACE1 needs to be further validated. In addition, Zeng T et al. have demonstrated that BACE1-AS is able to modulate BACE1 by serving as a competing endogenous RNA, and BACE1-AS blocks BACE1 mRNA degradation via the regulation of BACE1-targeting miRNAs [Citation23]. Moreover, it is suggested that BACE1-AS is upregulated in AD patients [Citation7], and miR-34b/c levels are downregulated in PD [Citation24], which are in line with the results in this current study.
Through numerous assays, we have also discovered that downregulation of BACE1-AS reduced MDA, Glu and iNOS levels and elevated SOD, GSH-Px, DA, and TH levels, and at the same time, repressed apoptosis and improved oxidative stress injury in the substantia nigra neurons of PD rats. It has been documented that in PD, the elevation of NOSusuallycorrelated with neuronal degeneration, and clinical symptoms of this disease are attributed to a decrease of midbrain DA neurons [Citation25,Citation26]. A study has revealed that the GSH-Px and SOD levels were substantiallydeclinedwhile the MDA content was elevated in PD model, which shows the involvement of oxidative stress injury in PD [Citation27]. The findings are directly in accordance with a previous finding indicating that silencing of BACE1-ASnonprotein-coding transcript regulates beta-amyloid-related hippocampal neurogenesis in AD [Citation28]. Zhang et al. confirmed that knockdown of BACE1-AS by siRNA improved memory and learning behaviors inaAD animal model [Citation29], and other lncRNAs have participated in PD. A recent study has revealed that silencing of lncRNA NEAT1 is capable of restricting MPTP-induced autophagy in vivo by stabilizing PINK1 protein and ameliorated dopaminergic neuronal injury in PD [Citation30]. A similar report by Lin Q et al. has shown that lncRNA HOTAIR boosts the development of PD by targeting miR-126-5p through RAB3IP [Citation31]. Furthermore, we revealed that knockdown of miR-34b-5p reversed the inhibitory impacts caused by the upregulation of lncRNA BACE1-AS on PD progression. There has been a literature suggesting that diminution of miR-34b and miR-34 c augments α-synuclein expression in PD, thereby catalyzing PD development [Citation12]. It was demonstrated that miR-34b/c controlled reactive oxygen species production and affected mitochondrial oxidative stress triggered by extremely low-frequency magnetic fields in neuronal cells, thereby regulating the risk of neurodegenerative diseases [Citation32]. The similar effects of other miRNAs have also been identified in PD. For instance, Dong RF et al. have demonstrated that mir-124-3p is likely to play a neuroprotective role in PD by regulating ANAX5 [Citation33]. An early finding has illustrated that elevation of miR-124 modulates apoptosis and autophagy process in MPTP-induced PD via targeting Bim, which further diminishing the decrease of dopaminergic neurons [Citation34]. All the above findings are consistent with what we have found in this study.
From the results, it is clear that downregulation of lncRNA BACE1-AS limits iNOS activation in the substantia nigra to alleviate dopamine-dependent oxidative stress injury in PD rats by upregulating miR-34b-5p and downregulating BACE1. Our study offers new clues for the role of the BACE1-AS/miR-34b-5p/BACE1 axis in rats with PD and emphasizes a new direction for the clinical treatment of PD. Nevertheless, further studies are still required for better elucidation of the specific mechanism of lncRNA BACE1-AS on PD.
Ethical statement
Animals were treated humanely in compliance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of Institute for Brain Sciences Research, School of Life Sciences, Henan University.
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this pap.
Disclosure statement
The authors declare that they have no conflicts of interest.
Additional information
Funding
References
- Schneider RB, Iourinets J, Richard IH. Parkinson’s disease psychosis: presentation, diagnosis and management. Neurodegener Dis Manag. 2017;7(6):365–376.
- Beitz JM. Parkinson’s disease: a review. Front Biosci (Schol Ed). 2014;6:65–74.
- Ascherio A, Schwarzschild MA. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol. 2016;15(12):1257–1272.
- Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386(9996):896–912.
- Li J, Meng H, Bai Y, et al. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23(5):205–217..
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166.
- Riva P, Ratti A, Venturin M. The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr Alzheimer Res. 2016;13(11):1219–1231.
- Liu S, Cui B, Dai Z-X, et al. Long non-coding RNA HOTAIR promotes Parkinson’s disease induced by MPTP through up-regulating the expression of LRRK2. Curr Neurovasc Res. 2016;13(2):115–120..
- Cao B, Wang T, Qu Q, et al. Long noncoding RNA SNHG1 promotes neuroinflammation in Parkinson’s disease via regulating miR-7/NLRP3 pathway. Neuroscience. 2018;388:118–127.
- Liu T, Liu Q, Zheng S, et al. MicroRNA-21 promotes cell growth and migration by targeting programmed cell death 4 gene in Kazakh’s esophageal squamous cell carcinoma. Dis Markers. 2014;2014:232837.
- Liu L, Liu L, Shi J, et al. MicroRNA-34b mediates hippocampal astrocyte apoptosis in a rat model of recurrent seizures. BMC Neurosci. 2016;17(1):56..
- Kabaria S, Choi DC, Chaudhuri AD, et al. Inhibition of miR-34b and miR-34c enhances alpha-synuclein expression in Parkinson’s disease. FEBS Lett. 2015;589(3):319–325..
- Feng T, Tammineni P, Agrawal C, et al. Autophagy-mediated regulation of BACE1 protein trafficking and degradation. J Biol Chem. 2017;292(5):1679–1690..
- Koelsch G. BACE1 function and inhibition: implications of intervention in the amyloid pathway of Alzheimer’s disease pathology. Molecules. 2017;22:10.
- Decressac M, Mattsson B, Bjorklund A. Comparison of the behavioural and histological characteristics of the 6-OHDA and alpha-synuclein rat models of Parkinson’s disease. Exp Neurol. 2012;235(1):306–315.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
- Coupland KG, Kim WS, Halliday GM, et al. Role of the long non-coding RNA MAPT-AS1 in regulation of microtubule associated protein Tau (MAPT) expression in Parkinson’s Disease. PLoS One. 2016;11(6):e0157924..
- Tarakad A, Jankovic J. Diagnosis and management of Parkinson’s Disease. Semin Neurol. 2017;37(2):118–126.
- Zhou Y, Gu C, Li J, et al. Aberrantly expressed long noncoding RNAs and genes in Parkinson’s disease. Neuropsychiatr Dis Treat. 2018;14:3219–3229.
- Zhang Y, Xing H, Guo S, et al. MicroRNA-135b has a neuroprotective role via targeting of beta-site APP-cleaving enzyme 1. Exp Ther Med. 2016;12(2):809–814..
- Gong G, An F, Wang Y, et al. miR-15b represses BACE1 expression in sporadic Alzheimer’s disease. Oncotarget. 2017;8(53):91551–91557..
- Fang M, Wang J, Zhang X, et al. The miR-124 regulates the expression of BACE1/beta-secretase correlated with cell death in Alzheimer’s disease. Toxicol Lett. 2012;209(1):94–105..
- Zeng T, Ni H, Yu Y, et al. BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs. J Chem Neuroanat. 2019;98:87–96.
- Hernandez-Rapp J, Rainone S, Hebert SS. MicroRNAs underlying memory deficits in neurodegenerative disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2017;73:79–86.
- Knott C, Stern G, Wilkin GP. Inflammatory regulators in Parkinson’s disease: iNOS, lipocortin-1, and cyclooxygenases-1 and −2. Mol Cell Neurosci. 2000;16(6):724–739.
- Stojkovska I, Wagner BM, Morrison BE. Parkinson’s disease and enhanced inflammatory response. Exp Biol Med (Maywood). 2015;240(11):1387–1395.
- Pan X, Chen C, Huang J, et al. Neuroprotective effect of combined therapy with hyperbaric oxygen and madopar on 6-hydroxydopamine-induced Parkinson’s disease in rats. Neurosci Lett. 2015;600:220–225.
- Modarresi F, Faghihi MA, Patel NS, et al. Knockdown of BACE1-AS nonprotein-coding transcript modulates beta-amyloid-related hippocampal neurogenesis. Int J Alzheimers Dis. 2011;2011:929042.
- Zhang W, Zhao H, Wu Q, et al. Knockdown of BACE1-AS by siRNA improves memory and learning behaviors in Alzheimer’s disease animal model. Exp Ther Med. 2018;16(3):2080–2086..
- Yan W, Chen Z-Y, Chen J-Q, et al. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem Biophys Res Commun. 2018;496(4):1019–1024..
- Lin Q, Hou S, Dai Y, et al. LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Biol Chem. 2018;400(9):1217–1228.
- Consales C, Cirotti C, Filomeni G, et al. Fifty-Hertz magnetic field affects the epigenetic modulation of the miR-34b/c in neuronal cells. Mol Neurobiol. 2018;55(7):5698–5714..
- Dong RF, Zhang B, Tai L-W, et al. The neuroprotective role of MiR-124-3p in a 6-hydroxydopamine-induced cell model of Parkinson’s disease via the regulation of ANAX5. J Cell Biochem. 2018;119(1):269–277..
- Wang H, Ye Y, Zhu Z, et al. MiR-124 regulates apoptosis and autophagy process in MPTP model of Parkinson’s disease by targeting to bim. Brain Pathol. 2016;26(2):167–176..
