ABSTRACT
The expression of a long noncoding RNA termed RUSC1-AS1 is dysregulated in breast cancer and laryngeal squamous cell carcinoma, and this dysregulation affects various tumor-associated biological processes. To our knowledge, the expression status and detailed roles of RUSC1-AS1 in cervical cancer as well as its regulatory mechanisms of action remain unknown. Therefore, the objectives of this study were to measure RUSC1-AS1 expression in cervical cancer, investigate the effects of RUSC1-AS1 on cervical cancer cells, and identify the mechanism underlying these effects. Herein, RUSC1-AS1 was found to be highly expressed in cervical cancer tissues and cell lines. High RUSC1-AS1 expression significantly correlated with the International Federation of Gynecology and Obstetrics (FIGO) stage, lymph node metastasis, and shorter overall survival among the patients with cervical cancer. Functional assays revealed that interference with RUSC1-AS1 expression suppressed cervical cancer cell proliferation, migration, and invasion in vitro; induced apoptosis in vitro; and impeded tumor growth in vivo. In addition, RUSC1-AS1 was demonstrated to act as a competing endogenous RNA of microRNA-744 (miR-744) and consequently increase B-cell lymphoma 2 (Bcl-2 or BCL2) expression levels in cervical cancer cells. Furthermore, either inhibition of miR-744 or restoration of Bcl-2 expression neutralized the effects of the RUSC1-AS1 silencing on the malignant characteristics of cervical cancer cells. Thus, RUSC1-AS1 promotes the aggressiveness of cervical cancer in vitro and in vivo by upregulating miR-744–Bcl-2 axis output. The RUSC1-AS1–miR-744–Bcl-2 pathway may be involved in cervical cancer pathogenesis and could serve as a novel target for anticancer therapies.
Introduction
Cervical cancer is the second most prevalent cancer among women and the fourth leading cause of gynecological-cancer–associated deaths globally [Citation1]. Approximately 530,000 new cases are expected and 275,000 deaths are caused by cervical cancer worldwide annually [Citation2]. Over 75% of these new cases and deaths occur in developing countries, including China [Citation3]. Multiple factors, including early sexual intercourse, an increased number of sexual partners, and persistent human papillomavirus infection, are reported to be involved in the pathogenesis of cervical cancer [Citation4,Citation5]. Nonetheless, the detailed mechanisms behind the malignant progression of cervical cancer remain largely unclear and need further elucidation. Despite considerable progress in the diagnostic and therapeutic approaches in recent years, the clinical outcomes of patients with cervical cancer unfortunately remain unsatisfactory [Citation6]. The 5-year survival rate of patients with cervical cancer diagnosed at advanced stages is less than 40%, which may be attributable to metastasis and tumor recurrence [Citation7,Citation8]. Therefore, elucidating the mechanisms underlying cervical carcinogenesis and cervical cancer progression is necessary and may facilitate the identification of novel therapeutic targets to improve the prognosis of patients with cervical cancer.
Noncoding RNAs have now become a hotspot in biomedical research [Citation9]. Long noncoding RNAs (lncRNAs) are a relatively new group of noncoding RNAs over 200 nucleotides long [Citation10]. LncRNAs contain no obvious open reading frame and therefore have no protein-coding ability [Citation11]. LncRNAs are proven to be key mediators of carcinogenesis and cancer progression [Citation12]. In particular, a wide range of lncRNAs is dysregulated in cervical cancer and is involved in the regulation of various cancer-related phenomena, including cell proliferation, the cell cycle, apoptosis, metastasis, angiogenesis, and chemoresistance [Citation13–Citation15]. The molecular mechanisms responsible for the actions of lncRNAs in human cancers are diverse and complex [Citation16,Citation17]. Accumulated evidence confirms the concept of competing endogenous RNA (ceRNA), which is an lncRNA that sponges a microRNA (miRNA, miR) to decrease the influence of this miRNA on its targets, thereby causing the upregulation of this miRNA’s target mRNAs [Citation18–Citation20].
MiRNAs are a subset of endogenous, single-stranded, noncoding small RNAs ranging in size from 17 to 21 nucleotides [Citation21]. MiRNAs regulate gene expression by interacting with a complementary sequence in the 3′-untranslated region (3′-UTR) of a target mRNA and by triggering translation suppression and/or mRNA degradation [Citation22]. One mRNA can be directly targeted by numerous miRNAs, whereas one miRNA is likely to target many mRNAs [Citation23]. MiRNAs are aberrantly expressed in nearly all human cancer types, including cervical cancer [Citation24–Citation26] and may function either as oncogenic miRNAs (oncomiRs) or tumor suppressor miRNAs depending on their target mRNAs [Citation27]. Hence, in-depth knowledge about novel lncRNAs and miRNAs involved in cervical cancer progression may contribute to the identification and validation of attractive therapeutic targets in this aggressive disease.
RUSC1-AS1 is dysregulated in breast cancer [Citation28] and laryngeal squamous cell carcinoma [Citation29], and this dysregulation is involved in the regulation of various tumor-associated biological processes. Nonetheless, to the best of our knowledge, the expression status and detailed roles of RUSC1-AS1 in cervical cancer as well as its regulatory mechanisms of action are still unknown. Therefore, the objectives of this study were to measure RUSC1-AS1 expression in cervical cancer, investigate the effects of RUSC1-AS1 on cervical cancer cells, and to identify the potential mechanism underlying these effects.
Materials and methods
Clinical tissue samples
The study protocol was approved by the Ethics Committee of Gaomi People’s Hospital and was performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from all the patients for the use of their clinical tissue samples. In total, 45 pairs of cervical cancer tissue samples and adjacent noncancerous tissue samples were collected from the patients with cervical cancer who underwent surgical resection in Gaomi People’s Hospital. None of the patients had received chemotherapy, radiotherapy, or other anticancer therapy prior to the surgical procedure. The tissue samples were snap-frozen in liquid nitrogen after the surgical resection and then transferred to a − 80°C freezer for storage until RNA extraction.
Cell lines and culture conditions
A normal human cervix epithelial cell line (Ect1/E6E7) was purchased from the American Type Culture Collection (Manassas, VA, USA). Four human cervical cancer cell lines (HeLa, C-33A, SiHa, and CaSki) were ordered from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) of heat-inactivated fetal bovine serum (FBS) and 1% (v/v) of a penicillin/streptomycin solution (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to culture all the above cell lines. All cells were grown at 37°C in an incubator supplied with 5% of CO2.
Oligonucleotide transfection
An miR-744 agomir (a chemically engineered oligonucleotide that upregulates miR-744, hereafter referred to as agomir-744), its negative control (NC) agomir (agomir-NC), an miR-744 antagomir (a chemically engineered oligonucleotide that downregulates miR-744, hereafter referred to as antagomir-744), and antagomir-NC were acquired from GenePharma Co., Ltd. (Shanghai, China). Small interfering RNA (siRNA) that was utilized to silence RUSC1-AS1 expression (si-RUSC1-AS1) and NC siRNA (si-NC) were chemically synthesized by RiboBio Co., Ltd. (Guangzhou, China). A Bcl-2–overexpressing plasmid (pCMV-Bcl-2) and the empty pCMV vector were generated by Generay Biotechnology Co., Ltd. (Songjiang, Shanghai, China). Cells were seeded in 6-well plates, and the aforementioned nucleic acids were transfected into the cells by means of the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA).
RNA extraction and reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
The TRIzol Reagent (Invitrogen) was employed to isolate total RNA from the tissue samples or from cells. The concentration of total RNA was measured on a NanoDrop spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). Total RNA was converted into cDNA using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The synthesized cDNA was used for the evaluation of miR-744 expression by means of the TaqMan MicroRNA qPCR Assay Kit (Applied Biosystems). The U6 small nuclear RNA served as an internal reference for miR-744 quantitation. To analyze Bcl-2 mRNA and RUSC1-AS1 expression, reverse transcription was conducted with the PrimeScript RT Reagent Kit (Takara Biotechnology Co., Ltd., Dalian, China). Next, qPCR was performed using SYBR Premix Ex Taq™ (Takara Biotechnology Co., Ltd.). Expression levels of Bcl-2 mRNA and RUSC1-AS1 were normalized to GAPDH expression. Relative gene expression was calculated by the 2–ΔΔCt method [Citation30].
Subcellular fractionation
The Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, Thorold, Canada) was employed for subcellular fractionation of cervical cancer cells.
A cell counting kit-8 (CCK-8) assay
Transfected cells were collected after 24 h of incubation and seeded separately in 96-well plates at a density of 3,000 cells/well. The cells were then incubated at 37°C and 5% CO2. The CCK-8 assay was performed at four time points: 0, 24, 48, and 72 h after inoculation. In particular, 10 µL of the CCK-8 stain (Beyotime Institute of Biotechnology, Shanghai, China) was added into each well, and the cells were incubated at 37°C and 5% CO2 for another 2 h. After that, a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA) was used to measure the optical density at 450 nm wavelength.
Flow-cytometric analysis for apoptosis detection
Transfected cells were treated with 0.25% trypsin (1×), rinsed with ice-cold phosphate-buffered saline (PBS) twice, and subjected to the measurement of the apoptotic rate using the Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (Biolegend, San Diego, CA, USA). The cells were then resuspended in 100 µl of binding buffer that was mixed with 5 µl of Annexin V-FITC and 5 µl of the propidium iodide solution that came with the kit. Following 20 min incubation at 37°C in darkness, the proportion (%) of apoptotic cells was assessed on a flow cytometer (FACScan™; BD Biosciences, Franklin Lakes, NJ, USA).
Migration and invasion assays
After 48 h of transfection, the invasive ability of cervical cancer cells was examined using 24-well Transwell chambers (24-well inserts; pore size: 8 µm; Corning Inc., Corning, NY, USA) precoated with Matrigel (BD Biosciences). Briefly, transfected cells were harvested and resuspended in an FBS-free culture medium. A total of 5 × 104 cells were seeded in the upper compartment of each insert, while the lower compartment was filled with 500 µl of DMEM containing 20% of FBS. After 24 h incubation, noninvading cells were gently removed by swabbing the top layer of Matrigel with a cotton swab. The invading cells adhering to the undersurface of the insert were fixed with 100% methanol followed by staining with 0.5% crystal violet and washing with PBS. The number of invading cells was determined in five randomly chosen visual fields per insert under a light microscope (×200 magnification; Olympus Corporation, Tokyo, Japan). The migratory capacity was determined by the same experimental procedures as in the invasion assay, except that the Transwell chambers were not precoated with Matrigel.
A subcutaneous heterotopic xenograft experiment
All animal experimental procedures were approved by the Animal Research Ethics Committee of Gaomi People’s Hospital and were performed in accordance with the Animal Protection Law of the People’s Republic of China-2009.
Lentiviruses expressing either a RUSC1-AS1–targeting short hairpin RNA (shRNA; sh-RUSC1-AS1) or an NC shRNA (sh-NC) were designed and packaged by GenePharma Co., Ltd. To obtain a stable RUSC1-AS1 knockdown cell line, the HeLa cells transfected with either sh-RUSC1-AS1 or sh-NC were selected with 2 μg/mL puromycin.
Female BALB/c nude mice (4–6 weeks old) were purchased from the Animal Center of the Second Military Medical University (Shanghai, China). Transplantation was conducted by subcutaneous inoculation of stably sh-RUSC1-AS1–transfected or sh-NC–transfected HeLa cells around the flanks of the mice. Each group contained three nude mice. Starting at 2 weeks after the subcutaneous implantation, the width and length of the tumor xenografts were measured every 2 days until day 28. Volumes of the tumor xenografts were calculated via the formula: Volume (mm3) = 0.5 × width2 (mm2) × length (mm). All the mice were euthanized on day 28. The subcutaneous tumor xenografts were excised and weighed.
Target prediction and a luciferase reporter gene assay
StarBase 3.0 (http://starbase.sysu.edu.cn/) was employed to analyze the interaction between the lncRNA and miRNA.
For the luciferase reporter assay, the fragments of RUSC1-AS1 containing either the wild-type (WT) miR-744–binding sequence or a mutant (MUT) miR-744–binding sequence were designed and chemically synthesized by GenePharma Co., Ltd., and were subcloned into the pmirGLO luciferase reporter vector (Promega Corporation, Madison, WI, USA), thus resulting in reporter plasmids RUSC1-AS1-WT and RUSC1-AS1-MUT, respectively. Cotransfection of the luciferase plasmid and either agomir-744 or agomir-NC into cells was performed using the Lipofectamine 2000 reagent. Luciferase activities were determined 48 h after the cell transfection via a Dual-Luciferase Reporter Assay System (Promega Corporation). The relative luciferase activity was normalized to the Renilla luciferase activity.
An RNA immunoprecipitation (RIP) assay
This assay was conducted by means of the EZ-Magna RIP RNA-binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). Cervical cancer cells were incubated with RIP lysis buffer. The whole-cell extracts were collected and subjected to overnight incubation at 4°C with magnetic beads conjugated with either an anti-Argonaute 2 (AGO2) or anti-immunoglobin G (IgG) antibody (Millipore). After the isolation of immunoprecipitated RNA, RT-qPCR was conducted to analyze the amounts and interaction of RUSC1-AS1 and miR-744 in cervical cancer cells.
Protein extraction and western blot analysis
Total protein was extracted from the transfected cells at 72 h after incubation using ice-cold radioimmunoprecipitation assay buffer (Invitrogen; Thermo Fisher Scientific, Inc.). The concentration of total protein was detected with the BCA Protein Assay Kit (Shanghai Qcbio Science and Technologies Co., Ltd., Shanghai, China). Equal amounts of protein samples were loaded onto each lane for SDS-PAGE, followed by transfer onto polyvinylidene difluoride membranes (Beyotime Institute of Biotechnology). After blockage at room temperature with 5% dried skimmed milk dissolved in Tris-buffered saline containing 0.1% of Tween 20 (TBST) for 1 h, the membranes were incubated with a rabbit anti-human Bcl-2 monoclonal antibody (1:1000 dilution; cat. # ab32124; Abcam, Cambridge, UK) or a rabbit anti-human GAPDH monoclonal antibody (1:1000 dilution; ab181603; Abcam) overnight at 4°C and next probed with a goat anti-rabbit IgG antibody conjugated with horseradish peroxidase (secondary antibody; 1:5000 dilution; ab205718; Abcam) at room temperature for 1 h. The protein signals were visualized with the Enhanced Chemiluminescence Kit (Pierce; Thermo Fisher Scientific, Inc.).
Statistical analysis
Data are presented as the mean ± standard deviation. Differences between two groups were analyzed by Student’s t test. One-way analysis of variance, followed by the Student–Newman–Keuls post hoc test, was performed for multigroup comparisons. The association between RUSC1-AS1 status and clinical variables among the patients was analyzed by the χ2 test. Survival curves were constructed by the Kaplan–Meier method, and differences in survival curves were examined by the logrank test. The correlation between RUSC1-AS1 and miR-744 expression levels was assessed via Spearman’s correlation analysis. P < 0.05 was assumed to indicate a statistically significant difference.
Results
RUSC1-AS1 expression is high in cervical cancer tissues and cell lines
To reveal the expression pattern of RUSC1-AS1 in cervical cancer, we assessed its expression in 45 pairs of cervical cancer tissue samples and adjacent noncancerous tissues by RT-qPCR. The results showed that RUSC1-AS1 expression was significantly higher in the cervical cancer tissue samples than in the noncancerous tissue samples (). RUSC1-AS1 expression was also quantified in four cervical cancer cell lines (HeLa, C-33A, SiHa, and CaSki) and a normal human cervix epithelial cell line (Ect1/E6E7) by the same method. The expression level of RUSC1-AS1 was considerably higher in all four cervical cancer cell lines than in Ect1/E6E7 cells ().
Figure 1. RUSC1-AS1 expression is high in cervical cancer tissue samples and cell lines. (a) RT-qPCR was performed to quantify RUSC1-AS1 expression in 45 pairs of cervical cancer tissue samples and adjacent noncancerous tissue samples. (b) The expression levels of RUSC1-AS1 in four cervical cancer cell lines (HeLa, C-33A, SiHa, and CaSki) and a normal human cervix epithelial cell line (Ect1/E6E7) were determined by RT-qPCR. (c) The correlation between RUSC1-AS1 expression status and overall survival of patients with cervical cancer was investigated by the Kaplan–Meier method and logrank test (P = 0.022). *P < 0.05 and **P < 0.01
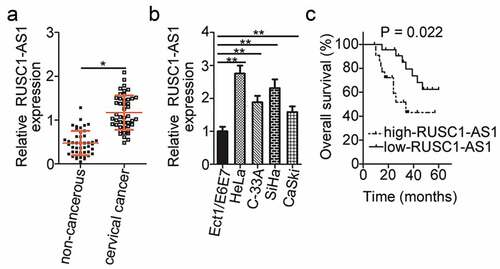
To evaluate the clinical significance of RUSC1-AS1 in cervical cancer, we subdivided all the patients with cervical cancer into either a low or high RUSC1-AS1 expression group based on the median value of RUSC1-AS1 expression among the cervical cancer tissue samples. Then, we investigated the association between RUSC1-AS1 expression and the clinical parameters among the patients with cervical cancer. Statistical analysis revealed that high RUSC1-AS1 expression notably correlated with the International Federation of Gynecology and Obstetrics (FIGO) stage (P = 0.017) and lymph node metastasis (P = 0.038) in patients with cervical cancer (). In addition, patients with cervical cancer in the high RUSC1-AS1 expression group showed shorter overall survival (, P = 0.022) as compared with the patients in the low RUSC1-AS1 expression group. These observations suggested that RUSC1-AS1 may play an important part in the initiation and progression of cervical cancer.
Table 1. The relation between RUSC1-AS1 expression and clinical variables of patients with cervical cancer
RUSC1-AS1 exerts oncogenic actions during cervical cancer progression
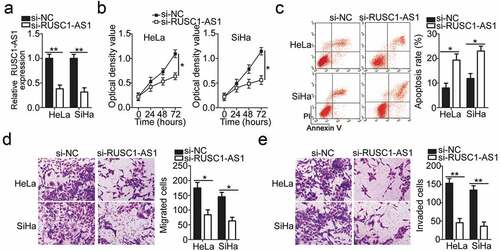
To illustrate the detailed involvement of RUSC1-AS1 in the progression of cervical cancer, cell lines HeLa and SiHa, which showed higher RUSC1-AS1 expression among the four cervical cancer cell lines, were chosen for functional analysis and were transfected with either si-RUSC1-AS1 or si-NC. In the two cell lines, the successful silencing of RUSC1-AS1 was confirmed by RT-qPCR (). The influence of the RUSC1-AS1 downregulation on the proliferation of cervical cancer cells was tested in the CCK-8 assay. The data revealed that the knockdown of RUSC1-AS1 decreased the proliferation of HeLa and SiHa cells (). In addition, we performed flow-cytometric analysis to determine whether the cell proliferation inhibition caused by the RUSC1-AS1 silencing was mediated by the promotion of apoptosis. The results indicated that the apoptosis rate of HeLa and SiHa cells was obviously higher in the si-RUSC1-AS1 group than in the si-NC group (). Furthermore, the migration and invasion assays were performed to examine the effects of RUSC1-AS1 on the migratory and invasive abilities of cervical cancer cells. Of note, transfection with si-RUSC1-AS1 resulted in a significant reduction of migration () and invasiveness () of HeLa and SiHa cells compared with the cells transfected with si-NC. These results suggested that RUSC1-AS1 has a cancer-promoting role in the growth and metastasis of cervical cancer cells in vitro.
Figure 2. RUSC1-AS1 knockdown inhibits HeLa and SiHa cell proliferation, migration, and invasion and promotes their apoptosis in vitro. (a) RUSC1-AS1 expression in HeLa and SiHa cells transfected with either si-RUSC1-AS1 or si-NC was measured via RT-qPCR. (b, c) The CCK-8 assay and flow-cytometric analysis were performed to quantitate the proliferation and apoptosis of HeLa and SiHa cells after transfection with either si-RUSC1-AS1 or si-NC. (d, e) Migration and invasion assays were conducted to analyze the effects of RUSC1-AS1 knockdown on the migration and invasiveness of HeLa and SiHa cells. *P < 0.05 and **P < 0.01
RUSC1-AS1 acts as a ceRNA and sponges miR-744 in cervical cancer cells
To elucidate the mechanisms by which RUSC1-AS1 performs its functions, we first analyzed the expression distribution of RUSC1-AS1 in HeLa and SiHa cells. The data indicated that RUSC1-AS1 was mainly localized in the cytoplasm of both HeLa and SiHa cells (), suggesting that this lncRNA may work as a ceRNA. Next, bioinformatics analysis was performed to search for an miRNA that can interact with RUSC1-AS1. MiR-744 was found to have a complementary binding site for RUSC1-AS1 (). MiR-744 was chosen for further experiments because this miRNA is implicated in the initiation and progression of cervical cancer [Citation31].
Figure 3. RUSC1-AS1 acts as a molecular sponge of miR-744 in cervical cancer cells. (a) The expression distribution of RUSC1-AS1 in HeLa and SiHa cells was studied by subcellular fractionation plus RT-qPCR analysis. (b) The wild-type (WT) and mutant (MUT) binding sites for miR-744 within RUSC1-AS1 as predicted by bioinformatics analysis. (c) HeLa and SiHa cells were transfected with either agomir-744 or agomir-NC. After that, RT-qPCR was carried out for determining the transfection efficiency. (d) The luciferase reporter assay was performed on HeLa and SiHa cells that were cotransfected with either agomir-744 or agomir-NC and the luciferase reporter plasmid carrying either the WT or mutant miR-744–binding site. (e) The RIP assay was conducted in HeLa and SiHa cell lysates, and the immunoprecipitated RNA was analyzed by RT-qPCR for assessing the enrichment of RUSC1-AS1 and miR-744 on AGO2-containing beads. (f) MiR-744 expression was measured by RT-qPCR in the 45 pairs of cervical cancer tissue samples and adjacent noncancerous tissue samples. (g) Spearman’s correlation analysis was carried out to evaluate the association between RUSC1-AS1 and miR-744 levels in cervical cancer tissues (r = – 0.5310; P = 0.0002). (h) Either si-RUSC1-AS1 or si-NC was introduced into HeLa and SiHa cells. MiR-744 expression in the transfected cells was evaluated via RT-qPCR. *P < 0.05 and **P < 0.01
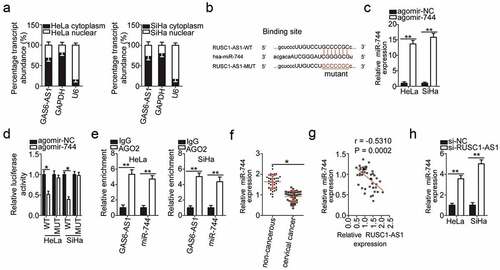
As presented in , transfection with agomir-744 dramatically increased the expression of miR-744 in HeLa and SiHa cells. Afterward, the luciferase reporter assay was applied to investigate whether miR-744 can directly bind to RUSC1-AS1 in cervical cancer cells. The results revealed that in HeLa and SiHa cells, exogenous miR-744 expression remarkably reduced the luciferase activity of the reporter plasmid harboring the wild-type miR-744–binding site (within the fragment of RUSC1-AS1); however, this effect was abrogated when the corresponding reporter plasmid with the mutant binding site was tested (). Furthermore, the RIP assay indicated that RUSC1-AS1 and miR-744 were enriched in the AGO2 complex in HeLa and SiHa cells (). Altogether, the luciferase reporter and RIP assays confirmed that miR-744 can directly bind to and interact with RUSC1-AS1 in cervical cancer cells.
The relation between RUSC1-AS1 and miR-744 was then examined by measuring miR-744 expression in the cervical cancer tissue samples. The expression of miR-744 was much lower in the cervical cancer tissue samples than in the matched noncancerous tissues (). An obvious inverse correlation was identified between RUSC1-AS1 and miR-744 levels in cervical cancer tissue samples by Spearman’s correlation analysis (; r = – 0.5310; P = 0.0002).
Finally, we determined the expression level of miR-744 in HeLa and SiHa cells after the knockdown of RUSC1-AS1. RT-qPCR analysis revealed that miR-744 was obviously upregulated by the depletion of RUSC1-AS1 in HeLa and SiHa cells (). Collectively, these results provided sufficient evidence that RUSC1-AS1 serves as a molecular sponge of miR-744 in cervical cancer cells.
BCL-2 expression is positively regulated by RUSC1-AS1 in cervical cancer cells
Bcl-2 (also known as BCL2) has been identified as a direct target gene of miR-744 in cervical cancer cells [Citation31]. Next, to test whether Bcl-2 expression can be modulated by RUSC1-AS1 in cervical cancer cells, the mRNA and protein levels of Bcl-2 were determined in HeLa and SiHa cells upon either si-RUSC1-AS1 or si-NC transfection. The mRNA () and protein () levels of Bcl-2 were both obviously lower in HeLa and SiHa cells after RUSC1-AS1 knockdown, as revealed by RT-qPCR and western blotting.
Figure 4. RUSC1-AS1 sponges miR-744 and consequently increases Bcl-2 expression in cervical cancer cells. (a, b) The mRNA and protein levels of Bcl-2 in HeLa and SiHa cells transfected with either si-RUSC1-AS1 or si-NC were measured by RT-qPCR and western blotting, respectively. (c) RT-qPCR was conducted to determine the efficiency of antagomir-744 transfection. (d, e) HeLa and SiHa cells were cotransfected with si-RUSC1-AS1 and either antagomir-744 or antagomir-NC. The amounts of Bcl-2 mRNA and protein were determined via RT-qPCR and western blotting, respectively. (f) Bcl-2 mRNA expression in the 45 pairs of cervical cancer tissue samples and adjacent noncancerous tissue samples was examined by RT-qPCR. (g) Spearman’s correlation analysis was performed to assess the relation between Bcl-2 mRNA and RUSC1-AS1 expression levels in cancerous tissues (r = 0.5262; P = 0.0005). *P < 0.05 and **P < 0.01
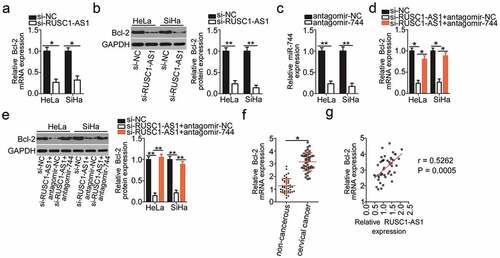
We next tested whether RUSC1-AS1 regulates the expression of Bcl-2 in cervical cancer through competitive binding with miR-744. To this end, the RUSC1-AS1–deficient HeLa and SiHa cells were next transfected with either antagomir-744 or antagomir-NC. First, RT-qPCR analysis validated the successful downregulation of miR-744 in HeLa and SiHa cells after the antagomir-744 transfection (). As expected, the Bcl-2 mRNA () and protein () levels were partially restored by the antagomir-744 transfection in the RUSC1-AS1–deficient HeLa and SiHa cells.
The expression in the Bcl-2 mRNA in the 45 pairs of cervical cancer tissue samples and adjacent noncancerous tissues was determined through RT-qPCR analysis. Bcl-2 mRNA turned out to be significantly upregulated in the cervical cancer tissues relative to the adjacent noncancerous tissues (). Notably, Spearman’s correlation analysis confirmed an obvious positive correlation between Bcl-2 mRNA and RUSC1-AS1 expression in the cancerous tissues (; r = 0.5262; P = 0.0005). Taken together, these data suggested that RUSC1-AS1 functioned as a ceRNA of miR-744 and consequently positively regulated Bcl-2 expression in cervical cancer cells.
RUSC1-AS1 promotes the aggressive phenotype of cervical cancer cells through the miR-744–Bcl-2 axis
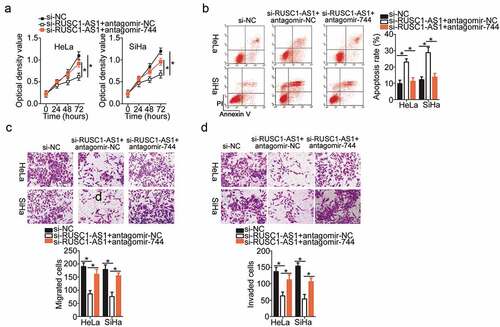
Rescue experiments were conducted to further study the participation of the miR-744–Bcl-2 axis in the mechanism of RUSC1-AS1’s actions on cervical cancer cells. First, rescue assays were carried out to validate the functional association between RUSC1-AS1 and miR-744 in cervical cancer cells. Si-RUSC1-AS1 and either antagomir-744 or antagomir-NC were cotransfected into HeLa and SiHa cells. Subsequently, we determined the impact of the antagomir-744 cotransfection on the malignant phenotype of si-RUSC1-AS1–transfected HeLa and SiHa cells. The miR-744 downregulation attenuated the effects of RUSC1-AS1 knockdown on HeLa and SiHa cell proliferation (), apoptosis (), migration (), and invasion ().
Figure 5. MiR-744 inhibition attenuated the actions of RUSC1-AS1 knockdown on the proliferation, apoptosis, migration, and invasiveness of cervical cancer cells. Either antagomir-744 or antagomir-NC was transfected into HeLa and SiHa cells in the presence of si-RUSC1-AS1. The transfected cells were subjected to the following assays. (a, b) The proliferation and apoptosis of the above-mentioned cells were determined in the CCK-8 and flow-cytometric assays. (c, d) The migratory and invasive capabilities of HeLa and SiHa cells treated as described above were evaluated in migration and invasion assays. *P < 0.05 and **P < 0.01
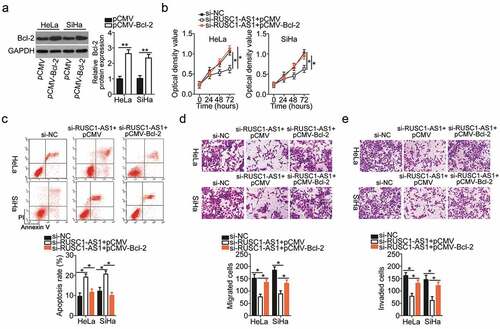
Next, the functional correlation between RUSC1-AS1 and Bcl-2 in cervical cancer cells was elucidated by means of rescue assays. The Bcl-2–overexpressing plasmid (pCMV-Bcl-2) or the empty vector (pCMV) was transiently transfected into the RUSC1-AS1 knockdown HeLa and SiHa cells. The transfection of pCMV-Bcl-2 into HeLa and SiHa cells efficiently increased the protein level of Bcl-2 (). Then, we subjected the HeLa and SiHa cells that were cotransfected with si-RUSC1-AS1 and either pCMV-Bcl-2 or pCMV to CCK-8, flow-cytometric, and migration and invasion assays. Restoration of Bcl-2 expression partially neutralized the influence of RUSC1-AS1 knockdown on the proliferation (), apoptosis (), migration (), and invasiveness () of HeLa and SiHa cells. In summary, these results implied that the tumor-promoting impact of RUSC1-AS1 on cervical cancer cells was mediated by the miR-744–Bcl-2 axis.
Figure 6. Restoration of the Bcl-2 level abrogates the impact of RUSC1-AS1 knockdown on the malignant phenotype of cervical cancer cells. (a) Bcl-2 protein expression was confirmed in the pCMV-Bcl-2–transfected or pCMV-transfected HeLa and SiHa cells by western blotting. (b–e) Si-RUSC1-AS1 was cotransfected with either pCMV-Bcl-2 or pCMV into HeLa and SiHa cells. Measurement of proliferation, apoptosis, migration, and invasiveness of the above-mentioned cells was performed by the CCK-8 assay, flow-cytometric analysis, and migration and invasion assays, respectively. *P < 0.05 and **P < 0.01
RUSC1-AS1 knockdown inhibits the xenograft growth of cervical cancer cells in vivo
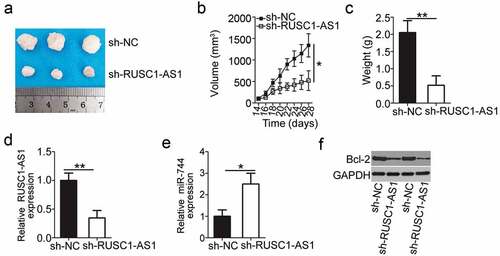
Finally, the subcutaneous heterotopic xenograft experiment was conducted to investigate the effect of RUSC1-AS1 knockdown on the tumor growth of cervical cancer cells in vivo. HeLa cells stably transduced with either the sh-RUSC1-AS1 lentivirus or the sh-NC lentivirus were injected subcutaneously into the flank of nude mice. Tumor-bearing mice were euthanized on day 28, and the tumor xenografts were excised and weighed. Compared with the sh-NC group, the mice injected with the sh-RUSC1-AS1–transfected HeLa cells developed obviously smaller tumor xenografts ( and b). The weight of tumor xenografts was significantly lower in the sh-RUSC1-AS1 group than in the sh-NC group (). The tumor xenografts were then processed for RT-qPCR and western blotting. RUSC1-AS1 expression was found to be decreased (), while miR-744 expression turned out to be upregulated () in the tumor xenografts derived from sh-RUSC1-AS1–transfected HeLa cells. Finally, western blotting was applied to measure Bcl-2 expression in the tumor xenografts, and the findings showed that the protein expression of Bcl-2 was lower in the tumor xenografts from the sh-RUSC1-AS1 group (). All these results indicated that the downregulation of RUSC1-AS1 retarded the tumor growth of cervical cancer cells in vivo by reducing miR-744–Bcl-2 axis output.
Figure 7. RUSC1-AS1 knockdown inhibits tumor growth of cervical cancer cells in vivo. (a) The subcutaneous tumor xenografts derived from sh-RUSC1-AS1–transfected or sh-NC–transfected HeLa cells on day 28 after inoculation. (b) The growth curve was plotted to monitor the volume of the tumor xenografts for 4 weeks. (c) Tumor weights in groups sh-RUSC1-AS1 and sh-NC were analyzed 4 weeks after the implantation. (d, e) Total RNA was isolated from the tumor xenografts and then used for evaluating RUSC1-AS1 and miR-744 expression by RT-qPCR. (e) Western blotting was conducted to assess Bcl-2 protein expression in the tumor xenografts. *P < 0.05 and **P < 0.01
Discussion
In recent decades, numerous lncRNAs have been reported to be aberrantly expressed in cervical cancer [Citation32–Citation34]. LncRNAs are believed to regulate a variety of cancer-related biological processes and to perform essential functions in the initiation and malignant progression of cervical cancer [Citation35–Citation37]. Hence, identification of the detailed functions of lncRNAs in cervical cancer and of their mechanisms of action is crucial for the discovery of promising targets for the diagnosis and therapy of patients with this malignant tumor. In this study, we evaluated RUSC1-AS1 expression in cervical cancer for the first time and determined the biological roles and the regulatory mechanism of RUSC1-AS1 action in cervical cancer.
RUSC1-AS1 expression is known to be increased in breast cancer [Citation28]. High RUSC1-AS1 expression is closely associated with tumor size and clinical grade of patients with breast cancer [Citation28]. Patients with breast cancer featuring high RUSC1-AS1 expression show poorer prognosis than do patients with low RUSC1-AS1 expression [Citation28]. Upregulation of RUSC1-AS1 is also observed in laryngeal squamous cell carcinoma [Citation29], and this upregulation negatively correlates with the patients’ overall survival [Citation29]. Nonetheless, the expression profile of RUSC1-AS1 in cervical cancer has remained largely unknown. In this study, RT-qPCR was carried out to quantify RUSC1-AS1 in cervical cancer tissues and cell lines. The results revealed that RUSC1-AS1 is significantly upregulated in cervical cancer tissues and cell lines. The high expression of RUSC1-AS1 significantly correlated with the FIGO stage and lymph node metastasis among the patients with cervical cancer. Notably, patients with cervical cancer in the high RUSC1-AS1 expression group showed shorter overall survival than did the patients in the low RUSC1-AS1 expression group. These findings suggest that RUSC1-AS1 is a potential biomarker for the diagnosis and prognosis of patients with cervical cancer.
RUSC1-AS1 acts as a pro-oncogenic lncRNA in carcinogenesis and cancer progression. For instance, silencing of RUSC1-AS1 obviously attenuates breast cancer cell growth, promotes cell cycle arrest, and induces apoptosis in vitro [Citation28]. Nonetheless, the specific functions of RUSC1-AS1 in cervical cancer have been unknown until our study. Here, functional experiments revealed that a knockdown of RUSC1-AS1 results in significant inhibition of cervical cancer cell growth, in apoptosis induction, and in cell migration and invasion suppression in vitro as well as an in vivo tumor growth slowdown.
The regulatory mechanisms by which lncRNAs exert their actions are complicated. One mode of action of lncRNAs is the ceRNA effect, where they can competitively interact with miRNAs, thereby upregulating miRNA target genes [Citation38]. The functions of lncRNAs are dependent on their subcellular localization. In this study, we first determined the expression distribution of RUSC1-AS1 in cervical cancer cells. RUSC1-AS1 turned out to be mainly localized in the cytoplasm of cervical cancer cells, suggesting that RUSC1-AS1 may serve as a ceRNA and be implicated in the regulation of a target protein’s expression at the post-transcriptional level.
The molecular events behind the oncogenic activity of RUSC1-AS1 in cervical cancer cells were further elucidated in detail. First, our bioinformatics analysis predicted that RUSC1-AS1 harbors a potential miR-744–binding site. Second, the luciferase reporter and RIP assay revealed that RUSC1-AS1 directly targets and interacts with miR-744 in cervical cancer cells. Third, we demonstrated that miR-744 expression is low in cervical cancer and inversely correlates with RUSC1-AS1 levels. Fourth, RUSC1-AS1 knockdown was found to increase endogenous miR-744 expression in cervical cancer cells. Fifth, the downregulation of RUSC1-AS1 decreased the expression of Bcl-2 in cervical cancer cells at both mRNA and protein levels, and these effects were mediated by the sponging of miR-744. Moreover, inhibition of miR-744 or restoration of Bcl-2 expression counteracted the effects of RUSC1-AS1 knockdown in cervical cancer cells. These findings collectively validate the interactions among Bcl-2 mRNA, miR-744, and RUSC1-AS1 in cervical cancer cells.
MiR-744 is known to be downregulated in cervical cancer [Citation31]. Exogenous miR-744 expression inhibits cervical cancer cell proliferation, colony-forming capacity, migration, and invasion in vitro and tumor growth in vivo [Citation31]. Regarding the mechanism, Bcl-2 mRNA has been demonstrated to be a direct target of miR-744 in cervical cancer [Citation31]. Bcl-2, an integral protein of the outer mitochondrial membrane, is strongly implicated in the initiation and malignant progression of cervical cancer by regulating cell proliferation, viability, cell cycle progression, apoptosis, epithelial–mesenchymal transition, migration, invasion, and metastasis [Citation39–Citation41]. Bcl-2 is reported as a double-edged sword by inducing apoptosis and inhibiting apoptosis at the same time [Citation42,Citation43]. Here, Bcl-2 mainly operate apoptosis induction activity. Another important finding of the present study is that the miR-744–Bcl-2 axis is essential for the biological actions of RUSC1-AS1 in cervical cancer cells. These results point to a cervical cancer pathogenesis–related regulatory network that is composed of RUSC1-AS1, miR-744, and Bcl-2. Hence, the RUSC1-AS1–miR-744–Bcl-2 pathway might be a promising therapeutic target in cervical cancer.
This study has two limitations. First, we did not use TCGA database to analyze the expression and clinical relevance of RUSC1-AS1 in cervical cancer. Second, the expression of Bcl-2 mRNA in cervical cancer was not examined using TCGA database. Also, the TCGA database was not applied to test the expression correlation between Bcl-2 mRNA and RUSC1-AS1. We will resolve the limitations in the near future.
Conclusions
In summary, RUSC1-AS1 promotes the malignancy of cervical cancer cells in vitro and in vivo by acting as a ceRNA on miR-744 and thereby increasing Bcl-2 expression. Our study provides functional evidence fully supporting the hypothesis that the RUSC1-AS1–miR-744–Bcl-2 pathway is an attractive target for the management of cervical cancer.
Disclosure statement
The authors declare that they have no competing interests.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
- Arbyn M, Castellsague X, de Sanjose S, et al. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686.
- Barra F, Lorusso D, Leone Roberti Maggiore U, et al. Investigational drugs for the treatment of cervical cancer. Expert Opin Invest Drugs. 2017;26:389–402.
- Bosch FX, de Sanjose S. Chapter 1: human papillomavirus and cervical cancer–burden and assessment of causality. J Nat Cancer Inst Monogr. 2003;3–13.
- Yu Y, Zhang Y, Zhang S. MicroRNA-92 regulates cervical tumorigenesis and its expression is upregulated by human papillomavirus-16 E6 in cervical cancer cells. Oncol Lett. 2013;6:468–474.
- Ghebre RG, Grover S, Xu MJ, et al. Cervical cancer control in HIV-infected women: past, present and future. Gynecol Oncol Rep. 2017;21:101–108.
- Smith RA, Brooks D, Cokkinides V, et al. Cancer screening in the United States, 2013: a review of current American Cancer Society guidelines, current issues in cancer screening, and new guidance on cervical cancer screening and lung cancer screening. CA Cancer J Clin. 2013;63:88–105.
- Kogo R, How C, Chaudary N, et al. The microRNA-218~Survivin axis regulates migration, invasion, and lymph node metastasis in cervical cancer. Oncotarget. 2015;6:1090–1100.
- Kanwal F, Lu C. A review on native and denaturing purification methods for non-coding RNA (ncRNA). J Chromatogr B Analyt Technol Biomed Life Sci. 2019;1120:71–79.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641.
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463.
- Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982.
- Tao L, Wang X, Zhou Q. Long noncoding RNA SNHG16 promotes the tumorigenicity of cervical cancer cells by recruiting transcriptional factor SPI1 to upregulate PARP9. Cell Biol Int. 2020 Mar;44(3):773–784. doi:10.1002/cbin.11272. Epub 2019 Dec 25.
- Duan W, Nian L, Qiao J, et al. LncRNA TUG1 aggravates the progression of cervical cancer by binding PUM2. Eur Rev Med Pharmacol Sci. 2019;23:8211–8218.
- Chang QQ, Chen CY, Chen Z, et al. LncRNA PVT1 promotes proliferation and invasion through enhancing Smad3 expression by sponging miR-140-5p in cervical cancer. Radiol Oncol. 2019;53:443–452.
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46.
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439.
- Huang MJ, Zhao JY, Xu JJ, et al. lncRNA ADAMTS9-AS2 Controls Human Mesenchymal Stem Cell Chondrogenic Differentiation and Functions as a ceRNA. Mol Ther Nucleic Acids. 2019;18:533–545.
- Dong XZ, Zhao ZR, Hu Y, et al. LncRNA COL1A1-014 is involved in the progression of gastric cancer via regulating CXCL12-CXCR4 axis. Gastric Cancer. 2020 Mar;23(2):260–272. doi:10.1002/cbin.11272. Epub 2019 Oct 24.
- Le K, Guo H, Zhang Q, et al. Gene and lncRNA co-expression network analysis reveals novel ceRNA network for triple-negative breast cancer. Sci Rep. 2019;9:15122.
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233.
- Zang H, Wang W, Fan S. The role of microRNAs in resistance to targeted treatments of non-small cell lung cancer. Cancer Chemother Pharmacol. 2017 Feb;79(2):227–231. doi:10.1007/s00280-016-3130-7. Epub 2016 Aug 11.
- Sharma N, Baruah MM. The microRNA signatures: aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol. 2018;21:126–144.
- Mandujano-Tinoco EA, Garcia-Venzor A, Melendez-Zajgla J, et al. New emerging roles of microRNAs in breast cancer. Breast Cancer Res Treat. 2018 Sep;171(2):247–259. doi:10.1007/s10549-018-4850-7. Epub 2018 Jun 9.
- Srivastava SK, Ahmad A, Zubair H, et al. MicroRNAs in gynecological cancers: small molecules with big implications. Cancer Lett. 2017;407:123–138.
- Xia H, Li Y, Lv X. MicroRNA-107 inhibits tumor growth and metastasis by targeting the BDNF-mediated PI3K/AKT pathway in human non-small lung cancer. Int J Oncol. 2016 Oct;49(4):1325–1333. doi:10.3892/ijo.2016.3628. Epub 2016 Jul 25.
- Hu CC, Liang YW, Hu JL, et al. LncRNA RUSC1-AS1 promotes the proliferation of breast cancer cells by epigenetic silence of KLF2 and CDKN1A. Eur Rev Med Pharmacol Sci. 2019;23:6602–6611.
- Hui L, Wang J, Zhang J, et al. lncRNA TMEM51-AS1 and RUSC1-AS1 function as ceRNAs for induction of laryngeal squamous cell carcinoma and prediction of prognosis. PeerJ. 2019;7:e7456.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408.
- Chen XF, Liu Y. MicroRNA-744 inhibited cervical cancer growth and progression through apoptosis induction by regulating Bcl-2. Biomed Pharmacothe. 2016;81:379–387.
- Hsu W, Liu L, Chen X, et al. LncRNA CASC11 promotes the cervical cancer progression by activating Wnt/beta-catenin signaling pathway. Biol Res. 2019;52:33.
- Song H, Liu Y, Jin X, et al. Long non-coding RNA LINC01535 promotes cervical cancer progression via targeting the miR-214/EZH2 feedback loop. J Cell Mol Med. 2019;23:6098–6111.
- Naghashi N, Ghorbian S. Clinical important dysregulation of long non-coding RNA CCHE1 and HULC in carcinogenesis of cervical cancer. Mol Biol Rep. 2019;46:5419–5424.
- Li YJ, Yang Z, Wang YY, et al. Long noncoding RNA ZNF667-AS1 reduces tumor invasion and metastasis in cervical cancer by counteracting microRNA-93-3p-dependent PEG3 downregulation. Mol Oncol. 2019;13:2375–2392.
- Yu CL, Xu XL, Yuan F. LINC00511 is associated with the malignant status and promotes cell proliferation and motility in cervical cancer. Biosci Rep. 2019 Sep 13;39(9). pii: BSR20190903. doi:10.1042/BSR20190903. Print 2019 Sep 30.
- Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909–4921.
- Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci. 2018 Apr 27;19(5). pii: E1310. doi: 10.3390/ijms19051310.
- Wang X, Xie Y, Wang J. Overexpression of MicroRNA-34a-5p Inhibits Proliferation and Promotes Apoptosis of Human Cervical Cancer Cells by Downregulation of Bcl-2. Oncol Res. 2018;26:977–985.
- Liu S, Wang H, Mu J, et al. MiRNA-211 triggers an autophagy-dependent apoptosis in cervical cancer cells: regulation of Bcl-2. Naunyn-Schmiedeberg’s Arch Pharmacol. 2020 Mar;393(3):359–370. doi:10.1007/s00210-019-01720-4. Epub 2019 Oct 21.
- Wang X, Yi Y, Lv Q, et al. Novel 1,3,5-triazine derivatives exert potent anti-cervical cancer effects by modulating Bax, Bcl2 and Caspases expression. Chem Biol Drug Des. 2018;91:728–734.
- Cleary ML, Smith SD, Sklar J. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell. 1986;47:19–28.
- Tsujimoto Y, Finger LR, Yunis J, et al. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097–1099.
