ABSTRACT
Long non-coding RNAs (lncRNAs) have been verified as a key modulator in tumor progression. However, the functions of lncRNAs in nasopharyngeal carcinoma (NPC) remain unclear. In the present study, we explored lncRNAs expression patterns in NPC tissues by GEO dataset and selected the high expression lncRNA (LINC01385) to further study. Our data showed that LINC01385 expression was significantly increased NPC and correlated with advanced clinical features and poor prognosis. Function assays showed that knockdown of LINC01385 expression reduced the proliferation and invasion abilities of NPC cells in vitro. In mechanism, LINC01385 acted as a molecular sponge of miR-140-3p in NPC cells, Twist1 mRNA was validated as a direct target of miR-140-3p in NPC cells. The effects of the LINC01385 knockdown on malignant characteristics of NPC cells were greatly attenuated by miR-140-3p inhibition or Twist1 overexpression. Thus, we illustrated that LINC01385 aggravated the progression of NPC by sponging miR-140-3p and upregulating Twist1 expression, which implied LINC01385 might serve as a new potential therapeutic target for NPC treatment.
Introduction
Nasopharyngeal carcinoma (NPC) is one of the major subtypes of head and neck cancers and exhibits a major cause of cancer-related death worldwide [Citation1,Citation2]. Despite novel improvements in NPC treatment in several aspects, the 5-year survival rate of NPC patients with distant metastases and drug resistance is still poor [Citation3,Citation4]. Therefore, it is an urgent need to identify the mechanism underlying NPC and develop new novel diagnostic and therapeutic strategies for the treatment of NPC.
Long non-coding RNA (lncRNA), a newly discovered type of RNA transcripts that are longer than 200 nucleotides (nt) without ability to encode protein [Citation5,Citation6]. Accumulating evidence demonstrated that lncRNAs play critical biological roles in diverse cellular processes, such as apoptosis, differentiation, invasion, and meiotic entry [Citation7–Citation9]. For example, Zhang et al found that MALAT1 was upregulated and promoted the proliferation and invasion of renal cancer cells [Citation10]. Chen et al showed that lncRNA CCAT1 promoted multiple myeloma cells proliferation through regulating the miR-181a-5p/HOXA1 axis [Citation11]. Gao et al found that lncRNA SBF2-AS1/miR-361-5p/FOXM1 axis could promote cervical cancer cells proliferation and invasion [Citation12]. However, the roles and underlying mechanisms of lncRNAs in tumorigenesis are still largely unclear.
MicroRNA (miRNA), a category of non-coding short RNA, could promote or inhibit cell progress through binding to target mRNAs at the 3ʹ-UTR [Citation13,Citation14]. Emerging evidence revealed that miR-140-3p was related to tumor progression. For example, Zhou et al showed that miR-140-3p was downregulated in breast cancer and inhibited proliferation and migration by targeting TRIM28 [Citation15]. Zhang et al revealed that miR-140-3p reduced hepatocellular carcinoma cells progression by inactivation of the MAPK axis [Citation16]. However, the roles and underlying mechanisms of miR-140-3p in NPC progression are still unclear.
In our study, we determined lncRNA expression patterns of NPC tissues by a published GEO dataset. Next, we explored the roles and underlying mechanisms of LINC01385 in NPC progression. Our results showed that LINC01385 was obviously increased and promoted NPC cells proliferation and invasion through regulating the miR-140-3p/Twist1 axis.
Materials and methods
Patient samples
NPC tissue samples and adjacent nontumor tissue samples were collected from 35 patients who underwent surgery at The First Affiliated Hospital of Zhengzhou University between June 2014 and June 2016. None of the patients had received chemotherapy or radiation therapy before surgery. Informed consents were obtained from all patients in a written format. This study was carried out with the approval of the Ethics Committees of The First Affiliated Hospital of Zhengzhou University and in accordance with the Declaration of Helsinki.
Cell culture and transfection
The human NPC cell lines (HNE1, 5–8 F, SUNE1, HK1) and nasopharyngeal epithelial cell line (NP69) were all provided by from Cell Bank of Type Culture Collection of Chinese Academy of Science (Shanghai, China). All cells were cultured as previous reports [Citation17,Citation18].
The small interfering RNA (siRNA) specific to LINC01385 (si-LINC01385), miR-140-3p mimics, miR-140-3p inhibitors and negative control (NC) were synthesized by RiboBio (Guangzhou, China). A plasmid encoding Twist1 (pcDNA3.1-Twist1) and the empty pcDNA3.1 vector were designed and constructed by GenePharma Technology (Shanghai, China). The transfections were carried out using Lipofectamine 3000 reagent (Invitrogen, CA, USA).
RNA extraction and quantitative real-time PCR
We extracted total RNA from cells and tissues using Trizol reagent (Invitrogen, CA, USA) and detected expressions of miRNA using Hairpin-itTM miRNAs qPCR kit (Genepharma, Shanghai). The U6 expression was used as a control. We detected LINC01385 and Twist1 expression through the SYBR green qPCR assay (Takara, Dalian). The GAPDH expression was used as a control. Relative gene expression was analyzed by the 2−ΔΔCT method.
Western blot assay
Cells were lysed in RIPA lysis Buffer containing PMSF (Sigma, MO, USA). 10% SDS-PAGE was employed to separate equal amounts of proteins and followed the proteins transferred PVDF membranes (Millipore, MA, USA). Subsequently, primary antibodies were used to incubate the membranes at 4°C overnight. Then the corresponding secondary antibody was used for 1 h at room temperature. Protein bands were quantified by ECL Kit (Bio-Rad, Hercules, CA, USA).
MTT assay
Transfected cells were inoculated into a 96-well plate (5 × 103 cells/well) in a total of 200 μl. To test cellular proliferation, 20 μl MTT solution was added to each well after indicated times. After 4 h, the medium was replaced by 150 μl DMSO. The absorbance at 450 nm wavelength was measured on a microplate reader (BioTek, VT, USA).
EdU assay
Cell proliferation was assessed by EdU assay kit (Ribobio, Guangzhou China). Transfected cells in 96-well plates were mixed with EdU medium diluent for 3 h, then with DAPI solution for nuclear staining. Proliferative cells were monitored by a fluorescent microscope (Leica, Wetzlar, Germany).
Colony formation assay
Cells were plated into 6-well plate at 500 cells per well for two weeks to form colonies. Crystal violet was used to stain colonies, and dissolved by 10% acetic acid, the colony was seen with naked eyes.
Wound healing assay
Cells were seeded in a 6-well culture plates to grow into a monolayer, and then scraped using 200uL pipette. The cells were further cultured in the medium for 24 h and closure of scratch was observed using a microscope at 40x magnification.
Transwell assay
Transwell chambers were coated with basement membrane Matrigel (BD Biosciences, CA, USA). 1 × 105 transfected cells were seeded into upper chamber with 200 μl serum-free medium. The lower chamber was supplemented with 750 μl medium containing 10% FBS. Following incubation for 24 h at 37°C, cells were fixed with 4% polyoxymethylene and stained with 0.5% crystal violet. Then stained cells were observed and counted under a microscope.
Dual luciferase reporter assay
The luciferase reporter vectors LINC01385-WT and Twist1-WT were formed via cloning the predictive miR-140-5p binding sites into the pmirGLO Dual-Luciferase Vector. The vectors LINC01385-Mut and Twist1-Mut were generated using point mutations of miR-140-3p binding sites. Cells were co-transfected with luciferase vectors and indicated transfection plasmids for 48 h, finally analyzed by Dual-Luciferase Reporter Assay System (Promega, Fitchburg, WI, USA).
Statistical analysis
Statistical analysis was performed by the SPSS21.0 software, and all data were presented as the mean ±SD. Comparison of the data between two groups was carried out by Student’s t test, whereas one-way analysis of variance along with Tukey’s post hoc test was performed to evaluate the statistical significance of differences among multiple groups. P < 0.05 expressed for a statistically significant difference.
Results
LINC01385 is a novel lncRNA in NPC progression
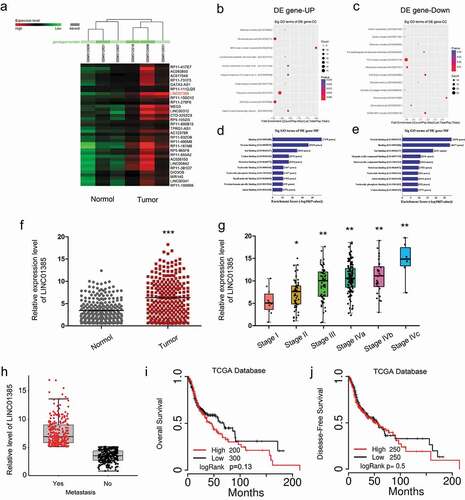
To explore the important lncRNAs involved in NPC progression, we first systematically analyzed the up-regulated lncRNAs in NPC tissues compared to adjacent normal tissues by using the online datasets (GSE12452), and LINC01385 is the most upregulated among all candidate lncRNA ()). Then, gene ontology (GO) analysis suggested that these differentially expressed lncRNAs are relevant to several vital physiological processes, and molecular functions (–)). Next, TCGA database results showed that LINC01385 expression was significantly increased NPC patients and associated with advanced tumor stage and metastasis (–)). Kaplan-Meier analysis revealed that high LINC01385 expression was significantly associated with poor overall survival (OS) and disease-free survival (DFS) in NPC patients (,)). These results demonstrated that LINC01385 might play critical roles in NPC tumorigenesis.
Figure 1. Dysregulated expression profiles of lncRNAs in NPC. (a) Aberrantly expression of lncRNAs was evaluated in GSE12452. (b, c) The top 10 upregulated and downregulated GO functions of the cellular component (CC) domain. (d, e) The top 10 upregulated and downregulated GO functions of the molecular function (MF) domains. (f) LINC01385 expression was increased in NPC tissues from TCGA dataset. (g, h) High LINC01385 expression was associated with advanced tumor stage and metastasis. (i, j) High LINC01385 expression was associated with poor OS and DFS in NPC patients. *P < 0.05.
LINC01385 is upregulated in NPC
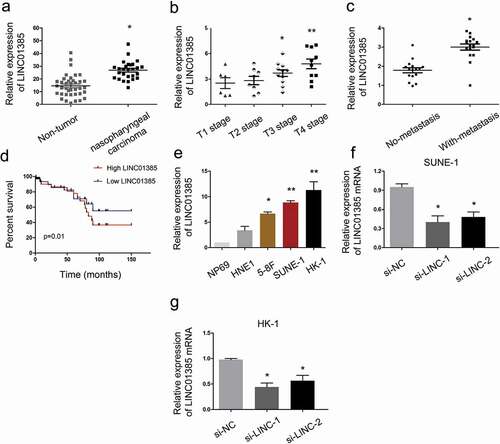
To assess the roles LINC01385 in NPC, 35 paired NPC tissues and adjacent non-tumor tissues specimens were collected. QRT-PCR showed that LINC01385 expression was significantly increased in NPC tissues compared to adjacent non-tumor tissues ()). High LINC01385 expression was associated with advanced tumor stage, metastasis and poor overall survival in NPC patients (–)). Subsequently, qRT-PCR showed that LINC01385 expression was increased in NPC cell lines compared with NP69 cells ()). Furthermore, si-LINC01385 was transfected into SUNE1 and HK-1 cells, which showed the highest LINC01385 expression among the four NPC cell lines (,)).
Figure 2. LINC01385 is upregulated in NPC. (a) LINC01385 expression was upregulated in NPC tissues. (b, c) High LINC01385 expression was associated with advanced tumor stage and metastasis. (d) LINC01385 expression was associated with poor OS in NPC patients. (e) LINC01385 was significantly increased in NPC cell lines compared with NP69 cells. (f, g) LINC01385 expression was measured in NPC cells transfected with si-LINC01385 or si-NC by qRT-PCR. *P < 0.05.
LINC01385 knockdown reduces NPC cells proliferation and invasion in vitro
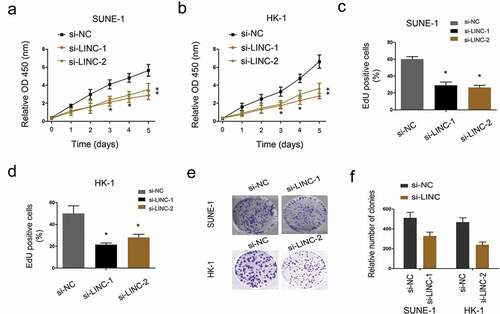
To evaluate the effects of LINC01385 on NPC cells viability, MTT and EdU assays were applied. Results showed that decreased LINC01385 expression notably attenuated the proliferation of SUNE1 and HK-1 cells (–)). Colony formation assay showed that LINC01385 inhibition significantly reduced NPC cell colony numbers (,)).
Figure 3. LINC01385 knockdown reduces NPC cells proliferation. (a, b) MTT assay was used to explore the proliferation of SUNE1 and HK-1 cells transfected with si-LINC01385 or si-NC. (c, d) EdU assay was used to determine SUNE1 and HK-1 cells viability in vitro. (e, f) Colony formation assays were explored to test the effects of si-LINC01385 on NPC cells colony formation ability. *P < 0.05.
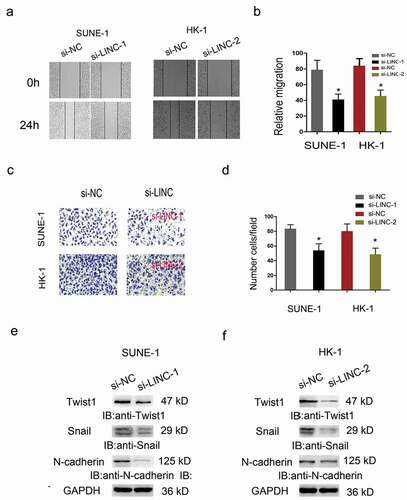
Furthermore, Wound healing and Transwell invasion assays were carried out to test the effects of LINC01385 on NPC cells metastasis. Results showed that the migration and invasion abilities in the si-LINC01385 group were significantly reduced compared to si-NC group (–)). The epithelial-mesenchymal transition (EMT) process plays an important role in tumor invasion and metastasis. Thus, we explored the effects of LINC01385 on EMT process, western blot showed that LINC01385 inhibition significantly reduced the expression of Twist1, Snail1, N-cadherin in NPC cells (,)).
Figure 4. LINC01385 knockdown reduced NPC cells metastasis in vitro. (a, b) Wound healing assay was used to explore the migration ability of SUNE1 and HK-1 cells transfected with si-LINC01385 or si-NC. (c, d) Transwell invasion assay was used to explore the invasion ability of SUNE1 and HK-1 cells transfected with si-LINC01385 or si-NC. (e, f) The effects of si-LINC01385 on Twist1, Snail1 and N-cadherin expression levels were determined by Western blot. *P < 0.05.
LINC01385 acts as a sponge for miR-140-3p
To further explore the underlying mechanism of LINC01385, bioinformatics prediction was used to predict the interaction between LINC01385 and miRNA, and miR-140-3p ranked top among all potential targets (–)). Next, we explored miR-140-3p expression in NPC tissues. QRT-PCR showed that low miR-140-3p expression was associated with advanced tumor stage, metastasis, and poor overall survival in NPC patients (–)). Subsequently, dual luciferase reporter assay showed that miR-140-3p mimics significantly reduced the luciferase activity of LINC01385-Wt group compared to LINC01385-Mut group ()). LINC01385 inhibition induced miR-140-3p expression in SUNE1 and HK-1 cells ()). Furthermore, correlation analysis showed that LINC01385 expression was negatively correlated with miR-140-3p expression in NPC tissues ()). These findings suggested that LINC01385 might act as a sponge for miR-140-3p in NPC.
Figure 5. LINC01385 acts as a sponge for miR-140-3p. (a, b) The information about LINC01385. (c) Venn diagrams showed the potential targets of LINC01385. (d) The predicted complementary binding sites between LINC01385 and miR-140-3p. (e-g) Low miR-140-3p expression was significantly associated with advanced tumor stage, metastasis, and poor overall survival rate in NPC patients. (h) Luciferase reporter assay for the luciferase activity in LINC01385-Wt and LINC01385Mut. (i) LINC01385 inhibition induced miR-140-3p expression in NPC cells. (j) LINC01385 expression was negatively correlated with miR-140-3p expression in NPC tissues. *P < 0.05.
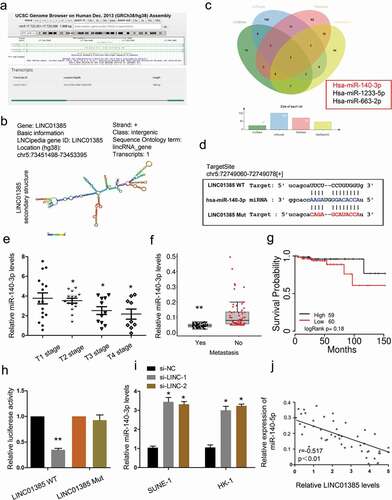
Twist1 is a target gene of miR-140-3p
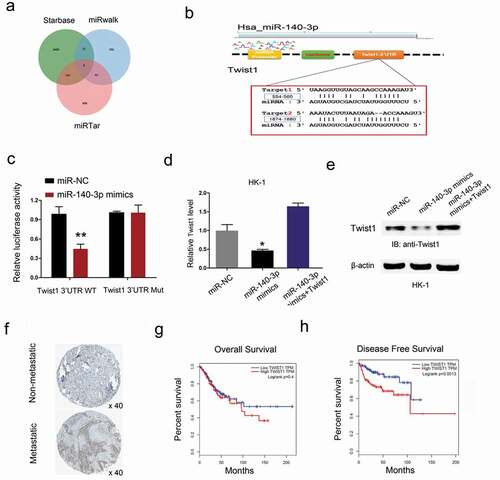
Next, the target gene of miR-140-3p was calculated by the Targetscan, miRwalk and miRTar. The results testified a target binding site between miR-140-3p and Twist1 (,)). Dual luciferase reporter assay showed miR-140-3p mimics reduced the luciferase activity of Twist1-Wt group compared to Twist1-Mut group ()). Results showed that miR-140-3p mimics significantly reduced Twist1 expression in HK-1 cells, while Twist1 overexpression abolished the effects (,)). Next, we explored Twist1 expression in NPC tissues. Results showed that Twist1 expression was upregulated in NPC patients with metastasis ()). Kaplan-Meier analysis showed that high Twist1 expression was associated with poor overall survival and disease-free survival in NPC patients (,)). These data suggested that Twist1 could act as a target of miR-140-3p in NPC.
Figure 6. Twist1 is a target gene of miR-140-3p. (a-b) Predicted binding sites for miR-140-3p in the 3ʹUTR of Twist1. (c) The relative luciferase activity was reduced in Twist1-Wt group compared to Twist1-Mut group. (d, e) miR-140-3p mimics significantly reduced Twist1 expression (mRNA and protein levels) in HK1 cells, while Twist1 overexpression abolished the effects. (f) Twist1 expression was upregulated in NPC patients with metastasis by IHC. (g, h) High Twist1 expression was associated with poor OS and DFS in NPC patients. *P < 0.05.
LINC01385 promotes NPC progression through miR-140-3p/Twist1 axis
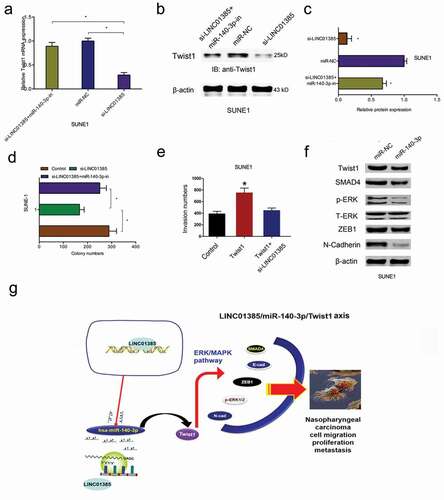
Next, we explored the LINC01385/miR-140-3p/Twist1 axis in NPC progression. We found that LINC01385 knockdown significantly reduced Twist1 expression both in mRNA and protein levels, while miR-140-3p inhibitors rescued the effects (–)). Colony formation assay showed that LINC01385 suppression reduced SUNE1 cells colony numbers, while the effects could be reversed by miR-140-3p inhibitors ()). Similarly, transwell assay showed that Twist1 overexpression promoted NPC cells invasion abilities, while LINC01385 inhibition abolished the effects ()). In addition, western blot showed that miR-140-3p mimics significantly reduced ERK/MAPK pathway-related gene expression (SMAD4, p-ERK, ZEB1, and N-cadherin) expression in SUNE1 cells ()). Thus, our data suggested that LINC01385 promoted the proliferation and metastasis of NPC by regulating miR-140-3p/Twist1/MAPK pathway ())
Figure 7. LINC01385 promotes NPC progression through miR-140-3p/Twist1 axis. (a-c) miR-140-3p inhibitors rescued the effects of LINC01385 knockdown on Twist1 expression both in mRNA and protein levels. (d) The anti-colony formation effects of LINC01385 suppression on SUNE1 cells could be abolished by miR-140-3p inhibitors. (e) Twist1 overexpression on SUNE1 cells invasion ability was rescued by LINC01385 suppression. (f) miR-140-3p mimics reduced ERK/MAPK pathway. (g) Schematic diagram of LINC01385 in NPC progression. *P < 0.05.
Discussion
Nasopharyngeal carcinoma (NPC) was a human malignancy tumor with atypical early symptoms to metastasis and the 5-year survival rate remains disappointing [Citation19]. Thus, it is necessary to find novel biomarkers and therapeutic targets for NPC early diagnosis and treatment. Recently, accumulating evidence suggested that lncRNAs play critical roles in NPC development. For example, Jia et al showed that lncRNA PXN-AS1-L promoted NPC progression through regulating the expression of SAPCD2 [Citation20]. Zhang et al suggested that lncRNA SOX2-OT promoted the growth and metastasis of NPC cells by regulating the miR-146b-5p/HNRNPA2B1 axis [Citation21]. Wen et al found that lncRNA DANCR stabilized HIF-1α and upregulated NPC metastasis ability through interacting with the NF90/NF45 complex [Citation22].
In our study, we identified a new lncRNA named LINC01385 which never been reported before. We confirmed that LINC01385 was significantly upregulated in NPC tissues and cell lines. High LINC01385 expression was associated with advanced tumor stage and metastasis in NPC patients. Kaplan-Meier analysis revealed that high LINC01385 expression was associated with poor overall survival rate and disease-free survival rate in NPC patients. In addition, we showed that silencing of LINC01385 restrained the proliferation and invasion of NPC cells in vitro. Thus, we demonstrated that LINC01385 might serve as an important regulator in NPC progression.
Accumulating studies showed that lncRNAs might act as miRNA sponges to influence the post transcription of miRNA [Citation23–Citation25]. In the present study, bioinformatics prediction showed that miR-140-3p was able to bind with LINC01385. And the results were further confirmed by dual-luciferase reporter assay. Subsequently, qRT-PCR showed that low miR-140-3p expression was significantly associated with advanced tumor stage, metastasis, and poor overall survival in NPC patients. Furthermore, rescue assay showed that the effects of LINC01385 suppression on SUNE-1 cells colony numbers could be reversed by miR-140-3p inhibitors. Thus, we suggested that LINC01385 might act as a miR-140-3p sponge in NPC progression.
Twist1, a member of the basic helix-loop-helix transcription factor family, is one of the master transcription factors that induce cell invasion and EMT during tumor progression [Citation26]. For example, Xu et al showed that Twist1 promoted the metastasis ability by reducing Foxa1 expression in breast cancer [Citation27]. Shen et al found that miR-203 reduced the proliferation of bladder cancer cells by targeting Twist1 [Citation28]. Li et al showed that lncRNA ATB promoted EMT process in breast cancer cells through regulating the miR-200 c/Twist1 axis [Citation29]. In our study, bioinformatic prediction and experimental verification showed that Twist1 could act as a direct target of miR-140-3p in NPC. High Twist1 expression was associated with metastasis and poor prognosis in NPC patients. Moreover, transwell assay showed that Twist1 overexpression promoted NPC cells invasion abilities, while LINC01385 inhibition abolished the effects. Therefore, we demonstrated that LINC01385 might promote NPC progression through targeting miR-140-3p/Twist1 pathway.
Conclusions
The present study demonstrated that LINC01385/miR-140-3p/Twist1 axis could promote cell proliferation, migration, invasion in NPC cells. Thus, we suggested that LINC01385 may be a potential therapeutic target in NPC treatment.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article.
Ethics approval and consent to participate
Ethical approval was given by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Acknowledgments
We are appreciating to every author for their assistance with the experiments and constructive comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012[J]. CA Cancer J Clin. 2015;65(2):87–108.
- Wei WI, Sham JST. Nasopharyngeal carcinoma[J]. Lancet. 2005;365(9476):2041–2054.
- Henle G, Henle W. Epstein‐Barr virus‐specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma[J]. Int J Cancer. 1976;17(1):1–7.
- Chua MLK, Wee JTS, Hui EP, et al. Nasopharyngeal carcinoma[J]. Lancet. 2016;387(10022):1012–1024.
- Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research?[J]. Oncogene. 2012;31(43):4577.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions[J]. Nat Rev Genet. 2009;10(3):155.
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development[J]. Nat Rev Genet. 2014;15(1):7.
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function[J]. Nat Rev Genet. 2016;17(1):47.
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts[J]. Nat Rev Mol Cell Biol. 2013;14(11):699.
- Zhang H, Yang F, Chen SJ, et al. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma[J]. Tumor Biol. 2015;36(4):2947–2955.
- Chen L, Hu N, Wang C, et al. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression[J]. Cell Cycle. 2018;17(3):319–329.
- Gao F, Feng J, Yao H, et al. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis[J]. Artif Cells Nanomed Biotechnol. 2019;47(1):776–782.
- Ambros V. The functions of animal microRNAs[J]. Nature. 2004;431(7006):350.
- Bartel DP. MicroRNAs: target recognition and regulatory functions[J]. cell. 2009;136(2):215–233.
- Zhou Y, Wang B, Wang Y, et al. miR-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28[J]. Oncol Lett. 2019;17(4):3835–3841.
- Zhang QY, Men CJ, Ding XW. Upregulation of microRNA‐140‐3p inhibits epithelial‐mesenchymal transition, invasion, and metastasis of hepatocellular carcinoma through inactivation of the MAPK signaling pathway by targeting GRN[J]. J Cell Biochem. 2019;120(9):14885–14898.
- Xie T, Pi G, Yang B, et al. Long non-coding RNA 520 is a negative prognostic biomarker and exhibits pro-oncogenic function in nasopharyngeal carcinoma carcinogenesis through regulation of miR-26b-3p/USP39 axis[J]. Gene. 2019;707:44–52.
- Hu X, Li Y, Kong D, et al. Long noncoding RNA CASC9 promotes LIN7A expression via miR‐758‐3p to facilitate the malignancy of ovarian cancer[J]. J Cell Physiol. 2019;234(7):10800–10808.
- Adham M, Kurniawan AN, Muhtadi AI, et al. Nasopharyngeal carcinoma in Indonesia: epidemiology, incidence, signs, and symptoms at presentation[J]. Chin J Cancer. 2012;31(4):185.
- Jia X, Niu P, Xie C, et al. Long noncoding RNA PXN‐AS1‐L promotes the malignancy of nasopharyngeal carcinoma cells via upregulation of SAPCD2[J]. Cancer Med. 2019. DOI:10.1002/cam4.2227
- Zhang E, Li X. LncRNA SOX2‐OT regulates proliferation and metastasis of nasopharyngeal carcinoma cells through miR‐146b‐5p/HNRNPA2B1 pathway[J]. J Cell Biochem. 2019;120(10):16575–16588.
- Wen X, Liu X, Mao YP, et al. Long non-coding RNA DANCR stabilizes HIF-1α and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma[J]. Theranostics. 2018;8(20):5676.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy[J]. Nat Rev Genet. 2016;17(5):272.
- Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development[J]. Biochimica Et Biophysica Acta (Bba)-gene Regulatory Mechanisms. 2016;1859(1):169–176.
- Ballantyne MD, McDonald RA, Baker AH. lncRNA/MicroRNA interactions in the vasculature[J]. Clin Pharmacol Ther. 2016;99(5):494–501.
- Qin Q, Xu Y, He T, et al. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms[J]. Cell Res. 2012;22(1):90.
- Xu Y, Qin L, Sun T, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression[J]. Oncogene. 2017;36(8):1157.
- Shen J, Zhang J, Xiao M, et al. miR-203 suppresses bladder cancer cell growth and targets Twist1[J]. Oncol Res Featuring Preclinical Clin Cancer Ther. 2018;26(8):1155–1165.
- Li RH, Chen M, Liu J, et al. Long noncoding RNA ATB promotes the epithelial− mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer[J]. Cell Death Dis. 2018;9(12):1171.
