ABSTRACT
Long non-coding RNA (lncRNA) SOX2 overlapping transcript (SOX2OT) has been shown to play an oncogenic role in diverse cancers, generating eight transcript variants. SOX2 is located in the third intron of SOX2OT. However, the biological function of SOX2OT in cervical cancer and implication with SOX2 remain to be further explored. In this study, we screened the expression pattern of different SOX2OT transcript variants in cervical cancer cells. Interestingly, both high-expression levels of SOX2OT transcript 7 (SOX2OT-7) and SOX2 were detected in C-33A (HPV−) and SiHa (HPV16+) cells. Thus, C-33A and SiHa cells were conducted to investigate the effects of SOX2OT on cell growth, migration and invasion. Finally, rescue experiments were performed to confirm the role of SOX2 in SOX2OT-mediated regulation of cervical cancer progression. The results showed that knockdown of SOX2OT suppressed cell viability, arrested cell cycle and ameliorated migration and invasion ability of C-33A and SiHa cells. Ectopic expression of SOX2OT-7 exacerbated cervical cancer cell proliferation, migration and invasion. In addition, we found that the expression levels and protein stability of SOX2 were positively regulated by SOX2OT. Inhibition of SOX2 could block the malignant phenotypes of C-33A and SiHa cells by SOX2OT-7. In conclusion, these findings indicate that lncRNA SOX2OT contributes to the growth, migration and invasion of cervical cancer cells by modulating SOX2. Importantly, we demonstrate that the transcript SOX2OT-7 may be a novel and promising biomarker for both HPV− and HPV16+ cervical cancer.
1. Introduction
Cervical cancer is one of the most prevalent gynecologic cancers in women worldwide with high mortality rate [Citation1,Citation2]. Human papillomavirus (HPV) has been identified as a major infective cause for the development of cervical cancer. Furthermore, numerous studies suggest a large proportion of cervical cancer patients are diagnosed as negative with HPV, which represents a poorer prognosis recently [Citation3–Citation5]. Thus, it is a significant importance for screening genes that implicated in both HPV-positive and -negative cervical cancer.
Sex-determining region Y-related high mobility group box 2 (SOX2) is a key transcription factor that involves in the pluripotency induction and maintenance of stem cells [Citation6]. Numerous reports have demonstrated the high expression of SOX2 in high-grade cervical intraepithelial neoplasia and cervical cancer tissues, indicating an independent poor prognosis [Citation7–Citation10]. SOX2 has been found to promote cell proliferation, migration, invasion and tumorigenicity of cervical cancer [Citation7,Citation11]. Furthermore, Liu et al. suggest that the endogenous SOX2 expression is co-regulated with the levels of stemness-related genes in cervical carcinomas [Citation12]. Therefore, it is beneficial for exploring a better understanding on the regulatory mechanism of SOX2 in cervical cancer.
Long non-coding RNAs (lncRNAs) are a prominent class of RNAs that control gene expression at epigenetic, transcriptional and posttranscriptional processes [Citation13]. SOX2OT overlapping transcript (SOX2OT), a conserved lncRNA located on human chromosome 3q26.3 (Chr3q26.3), which transcribes in the same orientation as SOX2 [Citation14]. Previous studies have demonstrated that overexpressing or silencing SOX2OT concordantly alters the expression of SOX2, suggesting the key correlations between SOX2OT and SOX2 [Citation15–Citation17]. It is reported that SOX2OT may encode different alternatively spliced transcript variants [Citation1–Citation8] to affect the phenotypes of various tumors [Citation15,Citation16,Citation18]. However, the role of SOX2OT in cervical cancer progression and its major isoform expression are unclear.
In this study, we first screened the expression profiles of SOX2OT variants and SOX2 in C-33A (HPV−), SiHa (HPV16+) and HeLa (HPV18+) cells using specific primers. Then, loss-of-function and gain-of-function studies were performed to test the effects of SOX2OT on cervical cancer cell proliferation, migration and invasion using C-33A and SiHa cells. Finally, specific siRNAs targeting SOX2 were conducted to discover the regulatory mechanism of SOX2OT.
2. Materials and methods
2.1. Cell culture and drug administration
Normal human epithelial cell line HaCaT and human cervical cancer cell line HeLa (HPV18+) were obtained from Procell (Wuhan, China). Another two human cervical cancer cell lines C-33A (HPV−) and SiHa (HPV16+) were obtained from Zhong Qiao Xin Zhou (Shanghai, China). Cells were cultured in minimum essential medium (MEM; 56416C, Sigma, St. Louis, MO, USA) containing 10% fetal bovine serum (FBS; F8067, Sigma) and incubated in a 5% CO2 incubator at 37°C. For the drug administration, at 48 h post-transfection, cells were treated with 5 μg/ml actinomycin D (MCE, New Jersey, USA) at 0, 0.5, 1.5, 3, 6, 12, 16, 20 h for further protein stability examination.
2.2. Cell transfection
To inhibit SOX2OT, two pairs of shRNAs targeting human SOX2OT (sh-SOX2OT #1 and sh-SOX2OT #2) and corresponding negative control (NC) shRNA were designed and synthesized by GenScript (Nanjing, China). Cells were seed into 6-well plates at a concentration of 2 × 105 cells per well. Twenty-four hours later, cells were transfected with sh-SOX2OTs or NC shRNA using Lipofectamine 2000 (11668–019, Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Furthermore, the amplified products of SOX2OT-7 were inserted into pcDNA3.1 plasmid vectors to construct SOX2OT-7 overexpressing plasmids. Then, SOX2OT-7 plasmids or control empty plasmids (Vector) were used to transfect into C-33A and SiHa cells by Lipofectamine 2000 following the manufacturer’s protocols. In addition, a pair of siRNAs targeting SOX2 (si-SOX2) and si-NC was purchased from JTS Scientific (Beijing, China). The cotransfection of SOX2OT-7 together with si-SOX2 or si-NC was also mediated by Lipofectamine 2000.
2.3. RT-PCR and quantitative real-time PCR (qRT-PCR)
Total RNAs were isolated from cells using TRIpure regent (RP1001, BioTeke, Beijing, China) and subsequently reverse-transcribed into complementary DNA (cDNA) using a Super-M-MLV Reverse Transcriptase kit (PR6502, BioTeke). RT-PCR reactions were performed to evaluate the expression levels of SOX2OT transcript variants and SOX2. PCR products were imaged on 1.5% agarose gel after electrophoresis. qRT-PCR was performed to determine the expressions of SOX2OT, SOX2OT-7 and SOX2 using SYBR Green Master Mix (SY1020, Solarbio, Beijing, China) by a qRT-PCR instrument (ExicyclerTM 96, Daejeon, Korea). The relative gene expression levels in C-33A and SiHa cells were calculated by the 2−ΔΔCT method. β-actin was used as an internal control. Specific sequences for RT-PCR and qRT-PCR primers () were manufactured by Sangon Biotech (Shanghai, China).
Table 1. The specific primer sequences for RT-PCR and qRT-PCR.
2.4. Cell proliferation assay
The viability of C-33A and SiHa cells was determined by Cell Counting kit-8 (CCK-8; C0037, Beyotime, Shanghai, China). In brief, cells were seeded in 96-well plates (4 × 103 per well) and then transfected with SOX2OT shRNAs or SOX2OT-7 overexpressing plasmids for 24 h, 48 h or 72 h. After incubation, cells were treated with CCK-8 reagent in each well and further cultured for 1 h. The absorbance at the wavelength of 450 nm was measured using a microplate reader (ELX-800, BIOTEK, Vermont, USA).
2.5. Flow cytometry analysis
The cell cycle distribution was examined using Cell Cycle Analysis kit (KGA512, KeyGEN, Nanjing, China). According to the standard protocol, cells were cultured for 48 h post transfection and then collected. Cells were fixed in ice-cold 70% ethanol for 2 h at 4°C and stained with propidium iodide/RNaseA solution at room temperature in dark for 60 min. Finally, cell cycle distribution was measured with the flow cytometer (C6, BD, New Jersey, USA).
2.6. Wound healing assay
C-33A and SiHa cells at 24 h post-transfection were used for wound healing assay to detect the cell migratory distance. Cells were incubated in serum-free medium and treated with mitomycin C (M0503, Sigma) for 1 h. A scratch of monolayer cells were made using a 200 μl pipette tip and then cultured for 0 h, 12 h and 24 h. The wound field was visualized by an inverted microscope (IX53, Olympus, Tokyo, Japan) at × 100 magnification, and the wound closing distance was calculated to assess the cell migration.
2.7. Transwell assay
For cell invasion assay, transwell inserts (3422, Corning Incorporated, Corning, NY, USA) were coated with Matrigel (356234, BD), and the suspensions were seeded into the upper chamber of Transwell inserts at a density of 1.5 × 104 cells per well. The lower chambers were filled with 30% FBS. After 24 h of culture, cells at the top surface of filters were removed by cotton swabs, while the invasive cells were fixed with 4% paraformaldehyde for 20 min and stained with 0.1% crystal violet (0528, Amresco, Ohio, USA) for 5 min. For cell migration assay, the similar experiments were performed, except the Transwell chambers without Matrigel coated. The stained cells were counted from five randomly selected fields in each well under an inverted microscope (IX53, Olympus) at × 200 magnification.
2.8. Western blot analysis
Total proteins were isolated using RIPA buffer (P0013B, Beyotime) containing PMSF (ST506, Beyotime) and quantified with a BCA Assay kit (P0009, Beyotime). Equal amount of each protein sample was loaded on the SDS-PAGE gel for electrophoresis, transferred onto PVDF membrane (IPVH00010, Millipore, Bedford, MA, USA) and subsequently blocked with 5% fresh skim milk for 1 h at room temperature. Then, the membranes were incubated with SOX2 antibody (1:1000; 20118-1-AP, Proteintech, Wuhan, China) and β-actin antibody (1:500; sc-47778, Santa, Dallas, TX, USA) at 4°C overnight. After washing in TBST, the membranes were incubated with the IgG-HRP goat anti-rabbit secondary antibody (1:5000; A0208, Beyotime) at 37°C for 45 min. The blots were visualized by ECL solution (P0018, Beyotime).
2.9. Statistical analysis
Statistics analyses from multiple groups were performed by one-way ANOVA, followed by Bonferroni’s post test with Graphpad Prism 7.0. Data were shown as mean ± SD. A p-value of less than 0.05 was considered statistically significant.
3. Results
3.1. Differential expression of SOX2OT transcripts and SOX2 in cervical cancer cells
To amplify the transcripts of SOX2OT, all primer sequences used are shown in . The expression levels of SOX2OT variants in cervical cancer cells were separated by agarose gel electrophoresis ()). The normal human epithelial cell line, HaCaT, a normal human epithelial cell line, was used as a control, which was found to only express SOX2OT variant 5 ()). Interestingly, a different expression profile of SOX2OT transcripts were observed in cervical cancer cells, in comparison to HaCaT cells. The transcripts of SOX2OT [Citation1,Citation4,Citation7,Citation8] were low-expressed in HeLa cells. We observed a significant high level of SOX2OT-7 and a low level of SOX2OT variant 1 in C-33A cells. Furthermore, only SOX2OT-7 was detected in SiHa cells. In addition, we noticed that no expression of SOX2OT variant 2, 3, 6 was detected in any cervical cancer cells. Notably, for the first time, we found the high expression of SOX2OT-7 in C-33A cells.
Table 2. The primer sequences for transcript variants and product length.
Figure 1. The expression profile of SOX2OT transcripts [Citation1–Citation8] and SOX2 in cervical cancer cell lines. (a) The transcripts of SOX2OT in cervical cancer cells were detected using Agarose gel electrophoresis. (b) The analysis of qRT-PCR and Agarose gel electrophoresis was used to detect the mRNA level of SOX2. (c) Western blot assay was performed to measure the protein level of SOX2.
![Figure 1. The expression profile of SOX2OT transcripts [Citation1–Citation8] and SOX2 in cervical cancer cell lines. (a) The transcripts of SOX2OT in cervical cancer cells were detected using Agarose gel electrophoresis. (b) The analysis of qRT-PCR and Agarose gel electrophoresis was used to detect the mRNA level of SOX2. (c) Western blot assay was performed to measure the protein level of SOX2.](/cms/asset/c225d00b-e0a8-47f9-8685-b54acb4e1878/kccy_a_1750812_f0001_b.gif)
SOX2 is shown to be associated with SOX2OT that served as a master regulator of pluripotency. We also examined the expression pattern of SOX2 mRNA and protein in different cervical cancers. As shown in ), SOX2 mRNA and protein were highly expressed in C-33A and SiHa cells. Considering that both C-33A and SiHa cells could express SOX2OT-7 and SOX2 from ), it might indicate that SOX2OT-7 expression was associated with SOX2 to regulate the development of HPV− and HPV16+ cervical cancer.
3.2. Knockdown of SOX2OT inhibited the growth, migration and invasion of C-33A and SiHa cells
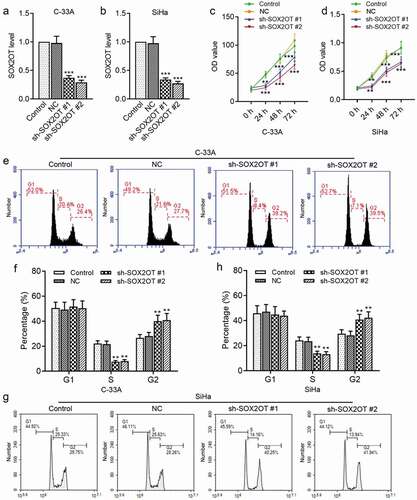
Then, we conducted specific shRNAs of SOX2OT to investigate its effects on cervical cancer malignancies. As indicated in ), SOX2OT shRNAs indeed induced a significant increase of SOX2OT expression level in C-33A and SiHa cells. It seemed from ) that the viable ratio of C-33A and SiHa cells was significantly reduced by SOX2OT silence. In addition, the distribution of cell cycle was also an important indicator for cell growth. We observed that knockdown of SOX2OT arrested the C-33A cell cycle at G2 phase by flow cytometry analysis ()). Furthermore, similar changes in the cell cycle distribution were also detected in SiHa cells ()).
Figure 2. SOX2OT knockdown suppressed the growth of C-33A and SiHa cells. (a–b) The expression level of SOX2OT in C-33A and SiHa cells was measured using qRT-PCR after transfected with SOX2OT shRNAs. (c–d) The viable cell number of C-33A and SiHa cells was detected by CCK-8 assay. (e–h) The analysis of flow cytometry was used to measure the cell cycle distribution of C-33A and SiHa cells. Results were expressed as mean ± SD. **p < 0.01, ***p < 0.001, compared to NC cells.
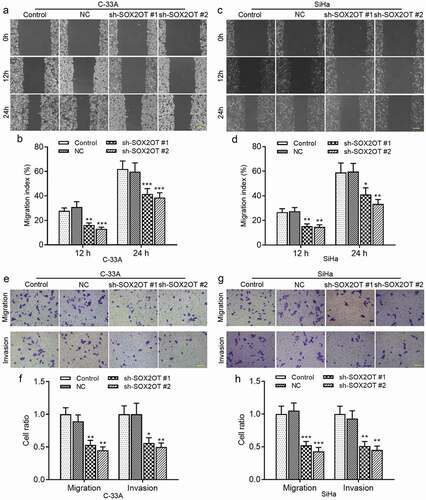
Next, we explored the role of SOX2OT in the migration and invasion of cervical cancer cells. As shown in ), wound healing assay indicated that the migratory distance of C-33A ()) and SiHa ()) cells was reduced by knockdown of SOX2OT. In addition, Transwell migration and invasion assay demonstrated that knockdown of SOX2OT reduced the number of migratory and invasive cells ()). Taken together, our results suggested that the inhibition of SOX2OT suppressed cell proliferation, migration and invasion of HPV− and HPV16+ cervical cancer.
Figure 3. SOX2OT knockdown inhibited the migration and invasion of C-33A and SiHa cells. (a–d) Wound healing assay was applied to detect the migratory distance of C-33A and SiHa cells. (e–g) The migration and invasion ability of C-33A and SiHa cells was measured by Transwell assay. Results were expressed as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, compared to NC cells.
3.3. Overexpression of SOX2OT-7 promoted the growth, migration and invasion of C-33A and SiHa cells
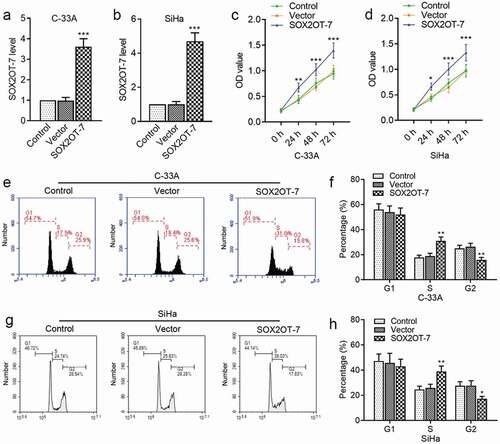
Considering that a novel splice variant SOX2OT-7 was highly expressed in C-33A and SiHa cells, the SOX2OT-7 overexpressing plasmids were constructed to further investigate its specific function. As expected, the level of SOX2OT-7 in C-33A ()) and SiHa ()) cells was significantly increased by the overexpressing plasmids. The results by CCK-8 assay suggested that SOX2OT-7 contributed to the survival rate of C-33A and SiHa cells ()). We also found that overexpression of SOX2OT-7 caused an accumulation of C-33A and SiHa cells at S phase and a decline at G2 phase ()).
Figure 4. SOX2OT-7 overexpression promoted the growth of C-33A and SiHa cells. (a–b) qRT-PCR analysis was used to determine SOX2OT-7 level in C-33A and SiHa cells transfected with SOX2OT-7 overexpressing plasmid. (c–d) The viability of C-33A and SiHa cells was detected using CCK-8 assay. (e–h) The cell cycle distribution of C-33A and SiHa cells was tested using flow cytometry analysis. Results were expressed as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, compared to Vector cells.
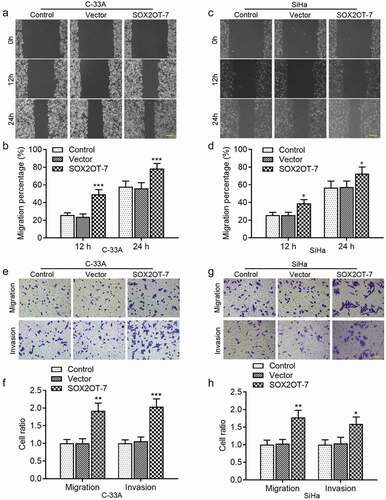
In addition, as illustrated in ), forced expression of endogenous SOX2OT-7 enhanced the mobility of C-33A and SiHa cells. The increments of migratory and invasive cells from the upper to lower chambers were also observed in ) using Transwell assay. These results revealed that SOX2OT-7 played an oncogenic role in the progression of both HPV− and HPV16+ cervical cancer.
Figure 5. SOX2OT-7 overexpression enhanced the migration and invasion of C-33A and SiHa cells. (a–d) The migratory distance of C-33A and SiHa cells was determined using wound healing assay. (e–h) The migration and invasion ability of C-33A and SiHa cells was measured using Transwell assay. Results were expressed as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, compared to Vector cells.
3.4. SOX2OT-7 positively regulated SOX2 expression in C-33A and SiHa cells
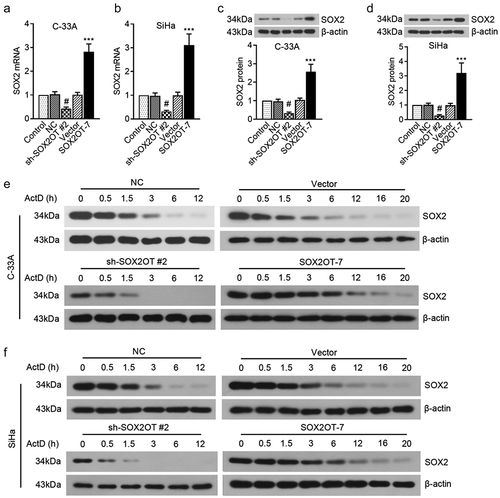
To investigate the underlying mechanism of SOX2OT on cervical cancer, we tested its effect on SOX2 expression. The results in ) demonstrated that both mRNA and protein expression levels of SOX2 in C-33A and SiHa cells were decreased by SOX2OT silence, whereas increased by SOX2OT-7 overexpression. Then the general transcription inhibitor, actinomycin D, was conducted to explore the role of SOX2OT-7 in SOX2 protein stability. As shown in ), the bands indicated that the degradation period of SOX2 protein in C-33A cells was shortened by silencing SOX2OT. However, overexpression of SOX2OT-7 significantly lengthened the degradation duration of SOX2 protein. Similar alterations were also observed in SiHa cells ()). Taken together, the results revealed that the novel transcript variant, SOX2OT-7, had positive associations with SOX2 expression and protein stability.
Figure 6. SOX2OT-7 positively regulated SOX2 expression in C-33A and SiHa cells. (a–b) The mRNA level of SOX2 in C-33A and SiHa cells transfected with sh-SOX2OT or SOX2OT-7 overexpressing plasmid was measured using qRT-PCR. (c–d) The protein expression of SOX2 in C-33A and SiHa cells transfected with sh-SOX2OT or SOX2OT-7 overexpressing plasmid was measured using Western blot. (e–f) Western blot assay was used to determine the degradation period of SOX2 protein in C-33A and SiHa cells after actinomycin D treatment. #p < 0.05, compared to NC cells; ***p < 0.001, compared to Vector cells. ActD: actinomycin D.
3.5. SOX2 was involved in the regulation of cervical cancer cell proliferation, migration and invasion by SOX2OT-7
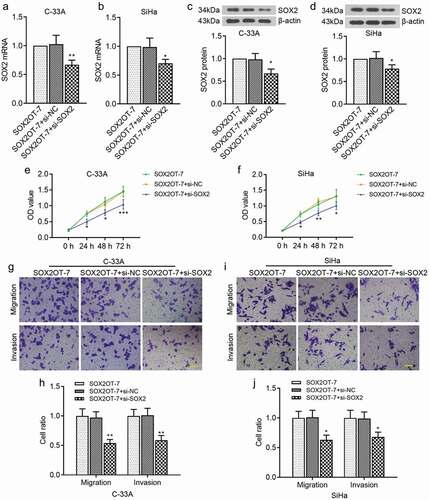
Considering the oncogenic role of SOX2OT-7 in cervical cancer and the close relationships between SOX2OT-7 and SOX2, we then focused on whether SOX2 was involved in the regulation of cervical cancer proliferation, migration and invasion by SOX2OT-7. SOX2OT overexpressing plasmid and si-SOX2 were cotransfected into C-33A and SiHa cells. It could be seen from ) that SOX2 mRNA and protein levels in C-33A and SiHa cells were expectantly decreased by si-SOX2. We found that inhibition of SOX2 could suppress the proliferation, migration and invasion of C-33A cells ()). Similarly, the malignant phenotypes of SiHa cells transfected with pcDAN-SOX2OT-7 were also rescued by SOX2 downregulation ()). Therefore, SOX2 mediated the regulation of cervical cancer proliferation, migration and invasion by SOX2OT-7.
Figure 7. SOX2 was involved in the regulation of cervical cancer cell proliferation, migration and invasion by SOX2OT-7. (a–b) qRT-PCR analysis was performed to measure SOX2 mRNA level in C-33A and SiHa cells. (c–d) Western blot assay was used to examine SOX2 protein level in C-33A and SiHa cells. (e–f) CCK-8 assay was applied to detect the viability of C-33A and SiHa cells. (g–j) Transwell assay was used to assess the migratory and invasive ability of C-33A and SiHa cells. *p < 0.05, **p < 0.01, ***p < 0.001, compared to SOX2OT-7+ si-NC cells.
4. Discussion
LncRNAs have been reported to be implicated in controlling the tumorigenesis and metastasis, which may be potential indicators in various cancer diagnosis or prognosis [Citation19]. Increasing studies suggested that high expression of lncRNA SOX2OT promoted cell proliferation, migration and invasion of various cancers, including esophagus, breast, hepatic and lung cancers [Citation15,Citation16,Citation18,Citation20]. Amaral et al. proposed that SOX2OT mediated the regulation of SOX2 at transcriptional level during the development of vertebrate [Citation21]. SOX2 is identified as an oncogene embedded in the third intron of SOX2OT, which transcribes in the same direction with SOX2OT. However, the role of SOX2OT and its association with SOX2 in cervical cancer have yet to be elusive. Here, a high expression of SOX2OT-7 and SOX2 was firstly observed in both C-33A and SiHa cells. Knockdown of SOX2OT could inhibit cervical cancer proliferation, cell cycle distribution, migration and invasion. Overexpression of SOX2OT-7 had opposite effects on cervical cancer development. In addition, SOX2 expression was positively regulated by SOX2OT, and inhibiting SOX2 impeded the effects of SOX2OT-7 on cervical cancer cell malignancies.
Previous studies suggested that Sox2ot was expressed similarly to Sox2 in mouse early differentiating embryoid bodies and neurospheres as well as some adult tissues, but it was also presented in tissues where Sox2 was not detected [Citation21], providing a clue for the functional importance of lncRNAs due to the cell type-specific regulation [Citation22]. Saghaeian Jazi and colleagues demonstrated that SOX2OT transcript variants [Citation1–Citation8] showed different expression profiles in different cell or tissue types [Citation23]. Therefore, in this study, we determined the specific expression patterns of SOX2OT variants in different cervical cancer cells using agarose gel electrophoresis. The results indicated that SOX2OT-7 was just expressed in SiHa cells, and significantly highly expressed in C-33A cells. Accumulating reports suggested that SOX2 expression was concordant with SOX2OT during the differentiation and carcinogenesis of stem cells, which was closely correlated with SOX2OT function [Citation15,Citation16,Citation18,Citation21]. We also found a high expression of SOX2 in both C-33A and SiHa cells. Thus, further experiments were focused on the effects of SOX2OT on HPV− and HPV16+ cervical cancer progression, especially SOX2OT-7.
The tumorigenesis and metastasis have been suggested to be critical biological processes of cancers. Hou et al. showed that SOX2OT contributed to lung cancer cell proliferation, and its high expression was correlated with poor survival [Citation18]. In non-small-cell lung cancer (NSCLC), the decrease of SOX2OT induced a suppression of cell migration and invasion by inhibiting epithelial–mesenchymal transition process [Citation24]. Consistent with these findings, our data demonstrated that inhibition of SOX2OT blocked cell proliferation, migration and invasion of cervical cancer. In addition, overexpression of SOX2OT transcript variants 4 and 7 caused facilitation for lung cancer cell in proliferation and migration [Citation17]. Wu and coworkers supported that the SOX2OT variant 4 promoted the proliferation of esophageal squamous cell carcinoma through the regulation of SOX2 [Citation25]. It was reported that SOX2OT-7 interacted with EGCG and Doxorubicin synergistically to inhibit osteosarcoma by autophagy and stemness inhibition [Citation26]. We found that overexpression of SOX2OT-7 exacerbated the aggressiveness of C-33A and SiHa cells. Altogether, the results suggested that SOX2OT functioned as an oncogene of cervical cancer, and the major isoform, SOX2OT-7, was important for HPV− and HPV16+ cervical cancer.
Increasing evidence suggested that SOX2 was over-expressed in multiple cancers, including cervical squamous cell carcinoma [Citation7,Citation19]. Dong et al. suggested that inhibition of SOX2 could suppress the proliferation, invasion and migration of glioblastoma cells [Citation27]. SOX2 was reported to be regulated by SOX2OT in breast cancer and esophageal squamous cell carcinoma [Citation16,Citation25]. However, the associations between SOX2OT and SOX2 in the regulation of cervical cancer were not well studied. Shahryari et al. demonstrated that SOX2OT variants 4, 5, 6 showed distinct expression patterns in parallel with NT2 cell differentiation, similar to that of SOX2 and OCT4 [Citation15]. SOX2OT-7 was over-expressed in NT2 cancer cell line with stem cell-like properties, coinciding with SOX2 gene [Citation23]. Consistent with these findings, our results showed that SOX2 expression levels were positively modulated by SOX2OT. Considering the nuclear localization of SOX2OT, it might be acted at transcriptional level rather than posttranscriptional level to regulate SOX2 expression from the literature by Shahryari et al. [Citation15]. In this experiment, we used actinomycin D (a transcriptional inhibitor) to investigate SOX2 stability. Knockdown of SOX2OT induced a quick degradation of SOX2 protein, whereas its overexpression prolonged the period of SOX2 degradation. Furthermore, the inhibition of SOX2 induced a suppression of malignant behaviors of C-33A and SiHa cells by SOX2OT-7. Thus, we concluded that SOX2OT-7 promoted cervical cancer cell proliferation, migration and invasion through the modulation of SOX2.
LncRNAs have been highlighted to modulate the biological function of miRNAs by acting as competing endogenous RNAs (ceRNAs) or molecular sponges [Citation28]. Previous studies suggested that multiple miRNAs participated in the regulation of cancer malignancies, such as miR-132, miR-200, miR-375 and miR-211 [Citation19,Citation24,Citation29–Citation31]. Therefore, we speculated that a ceRNA regulatory network might be associated with SOX2OT-7 in cervical cancer, which will be a probable interest in future work.
5. Conclusions
In summary, this present study suggests that lncRNA SOX2OT functions as an oncogenic gene in cell growth, migration and invasion of HPV− and HPV16+ cervical cancer through the modulation of SOX2. Especially, we first put forward that SOX2OT-7, one of the SOX2OT transcript variants, may be an important and probable biomarker in both HPV− and HPV16+ cervical cancers.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Egawa N, Egawa K, Griffin H, et al. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses. 2015;7(7):3863–3890.
- Ogilvie GS, van Niekerk D, Krajden M, et al. Effect of screening with primary cervical HPV testing vs cytology testing on high-grade cervical intraepithelial neoplasia at 48 months: the HPV FOCAL randomized clinical trial. Jama. 2018;320(1):43–52. .
- Riou G, Favre M, Jeannel D, et al. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA. Lancet. 1990;335(8699):1171–1174.
- Rodriguez-Carunchio L, Soveral I, Steenbergen RD, et al. HPV-negative carcinoma of the uterine cervix: a distinct type of cervical cancer with poor prognosis. Bjog. 2015;122(1):119–127.
- Nicolas I, Marimon L, Barnadas E, et al. HPV-negative tumors of the uterine cervix. Mod Pathol. 2019;32(8):1189–1196. .
- Liu K, Lin B, Zhao M, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013;25(5):1264–1271.
- Chang X, Zhang J, Huang C, et al. Sex-determining region Y-related high mobility group box (SOX)-2 is overexpressed in cervical squamous cell carcinoma and contributes cervical cancer cell migration and invasion in vitro. Tumour Biol. 2015;36(10):7725–7733.
- Canham M, Charsou C, Stewart J, et al. Increased cycling cell numbers and stem cell associated proteins as potential biomarkers for high grade human papillomavirus+ve pre-neoplastic cervical disease. PLoS One. 2014;9(12):e115379. .
- Stewart CJR, Crook ML:. Podoplanin and SOX2 Expression in CIN 3-like squamous cell carcinoma of the cervix. Int J Gynecol Pathol. 2018;37(1):59–67.
- Hou T, Zhang W, Tong C, et al. Putative stem cell markers in cervical squamous cell carcinoma are correlated with poor clinical outcome. BMC Cancer. 2015;15:785.
- Ji J, Zheng PS:. Expression of Sox2 in human cervical carcinogenesis. Hum Pathol. 2010;41(10):1438–1447.
- Liu XF, Yang WT, Xu R, et al. Cervical cancer cells with positive Sox2 expression exhibit the properties of cancer stem cells. PLoS One. 2014;9(1):e87092.
- Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24(11):651–663.
- Shahryari A, Jazi MS, Samaei NM, et al. Long non-coding RNA SOX2OT: expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front Genet. 2015;6:196.
- Shahryari A, Rafiee MR, Fouani Y, et al. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells. 2014;32(1):126–134.
- Askarian-Amiri ME, Seyfoddin V, Smart CE, et al. Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS One. 2014;9(7):e102140.
- Saghaeian Jazi M, Samaei NM, Ghanei M, et al. Overexpression of the non-coding SOX2OT variants 4 and 7 in lung tumors suggests an oncogenic role in lung cancer. Tumour Biol. 2016;37(8):10329–10338.
- Hou Z, Zhao W, Zhou J, et al. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol. 2014;53:380–388.
- Zhou K, Zhang C, Yao H, et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17(1):105.
- Shi XM, Teng F. Up-regulation of long non-coding RNA Sox2ot promotes hepatocellular carcinoma cell metastasis and correlates with poor prognosis. Int J Clin Exp Pathol. 2015;8(4):4008–4014.
- Amaral PP, Neyt C, Wilkins SJ, et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15(11):2013–2027.
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec(1):R17–29.
- Saghaeian Jazi M, Samaei NM, Ghanei M, et al. Identification of new SOX2OT transcript variants highly expressed in human cancer cell lines and down regulated in stem cell differentiation. Mol Biol Rep. 2016;43(2):65–72.
- Zhang K, Li Y, Qu L, et al. Long noncoding RNA Sox2 overlapping transcript (SOX2OT) promotes non-small-cell lung cancer migration and invasion via sponging microRNA 132 (miR-132). Onco Targets Ther. 2018;11:5269–5278.
- Wu Y, Chen X, Liang Y, et al. Overexpression of long non-coding RNA SOX2OT promotes esophageal squamous cell carcinoma growth. Cancer Cell Int. 2018;18:76.
- Wang W, Chen D, Zhu K. SOX2OT variant 7 contributes to the synergistic interaction between EGCG and Doxorubicin to kill osteosarcoma via autophagy and stemness inhibition. J Exp Clin Cancer Res. 2018;37(1):37.
- Dong H, Hao X, Cui B, et al. MiR-429 suppresses glioblastoma multiforme by targeting SOX2. Cell Biochem Funct. 2017;35(5):260–268.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358.
- Li Z, Jiang P, Li J, et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. 2018;37(28):3822–3838. .
- Shafiee M, Aleyasin SA, Mowla SJ, et al. The effect of MicroRNA-375 overexpression, an inhibitor of helicobacter pylori-induced carcinogenesis, on lncRNA SOX2OT. Jundishapur J Microbiol. 2016;9(9):e23464.
- Shafiee M, Aleyasin SA, Vasei M, et al. Down-regulatory effects of miR-211 on long non-coding RNA SOX2OT and SOX2 genes in esophageal squamous cell carcinoma. Cell J. 2016;17(4):593–600.
